
Deinococcus koreensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
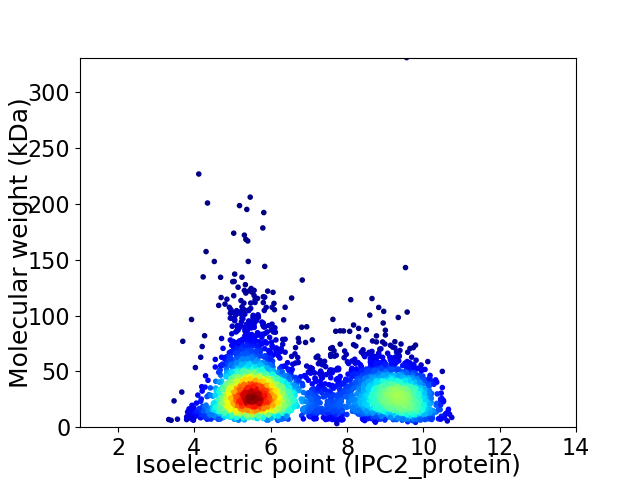
Virtual 2D-PAGE plot for 3967 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K3UYP5|A0A2K3UYP5_9DEIO Restriction endonuclease subunit R OS=Deinococcus koreensis OX=2054903 GN=CVO96_09995 PE=4 SV=1
MM1 pKa = 6.91NTTPRR6 pKa = 11.84VLALMAALAAGTAAAATTNTTAGQIITNQATATFTDD42 pKa = 4.15PTTLAAATPIVSNTVQTVVLPKK64 pKa = 10.18PGFDD68 pKa = 2.85IQYY71 pKa = 10.81ADD73 pKa = 3.72GSADD77 pKa = 3.38NTTATAPAPSFDD89 pKa = 3.64KK90 pKa = 11.04TGVLPGTTVTTGYY103 pKa = 10.84VVVNTGNVNGYY114 pKa = 8.52VVNLAADD121 pKa = 3.84STGGNAPQDD130 pKa = 3.41VKK132 pKa = 11.58YY133 pKa = 11.1YY134 pKa = 11.08LDD136 pKa = 4.05ANNDD140 pKa = 3.72GVPDD144 pKa = 3.49SSTPVTSVTLNADD157 pKa = 3.86NPNTPADD164 pKa = 3.75EE165 pKa = 5.11GIVRR169 pKa = 11.84ILQVITVASTAGTGAQYY186 pKa = 10.78SASPAGTAPSGSVSATDD203 pKa = 3.29TSTGITTAYY212 pKa = 9.22PYY214 pKa = 11.04AALTEE219 pKa = 4.36AQANPASTNGDD230 pKa = 3.41LQYY233 pKa = 11.15TRR235 pKa = 11.84VTVFTPTVTNAPADD249 pKa = 4.15GDD251 pKa = 4.07PATAGQQTPTTTVVVPPSASTVIDD275 pKa = 3.89PNNPTSAPGTPSSPSDD291 pKa = 3.42PTQPGYY297 pKa = 9.62TDD299 pKa = 3.81PPVAGSTGAVAVTVVGNVQTAYY321 pKa = 10.12PPADD325 pKa = 3.39TNAAADD331 pKa = 3.56AVNFKK336 pKa = 9.38NTVTTPAGSPADD348 pKa = 3.94TVNLFPVNPALNVGDD363 pKa = 4.23AGYY366 pKa = 9.83GAPYY370 pKa = 8.48GTNNGDD376 pKa = 3.84GSFTLPDD383 pKa = 3.4GTVVRR388 pKa = 11.84FLNADD393 pKa = 3.56GSAPTLVTSPVDD405 pKa = 3.38GKK407 pKa = 10.73AYY409 pKa = 9.64PVVSVPAGGATVSYY423 pKa = 8.55ITQVVYY429 pKa = 10.45PDD431 pKa = 4.22SNSLTNPTPITVVVGADD448 pKa = 3.05SGNDD452 pKa = 3.25YY453 pKa = 11.48NLIADD458 pKa = 4.39GTTTDD463 pKa = 3.78KK464 pKa = 11.16VLPPALQFGDD474 pKa = 3.93SNGVQTPPTADD485 pKa = 3.51ASAQPSEE492 pKa = 4.42TVTPGAAVSTGTPTPGMTTDD512 pKa = 3.45SSAVFPMDD520 pKa = 2.76IANPGEE526 pKa = 4.16YY527 pKa = 10.27GDD529 pKa = 4.62TYY531 pKa = 10.21TLSGSVLVPLSNGTTATVPVKK552 pKa = 10.78YY553 pKa = 10.28VDD555 pKa = 3.31ASGTEE560 pKa = 4.09LPKK563 pKa = 10.81NAAGQYY569 pKa = 6.33ITPVVDD575 pKa = 4.63ANAEE579 pKa = 4.0YY580 pKa = 10.12RR581 pKa = 11.84VYY583 pKa = 11.0AVVDD587 pKa = 3.49IPSNARR593 pKa = 11.84LTLPGSPLSVQQTVTSNYY611 pKa = 10.8SNITLQDD618 pKa = 3.46TNDD621 pKa = 4.83LIRR624 pKa = 11.84VGAIGGITVDD634 pKa = 3.48KK635 pKa = 9.49YY636 pKa = 10.47QAVGAAPSQTAAGKK650 pKa = 8.12TIKK653 pKa = 9.4TALPGEE659 pKa = 4.48TINYY663 pKa = 9.52AIVARR668 pKa = 11.84NSYY671 pKa = 10.7NDD673 pKa = 3.29AVKK676 pKa = 10.87NFVLSDD682 pKa = 3.37VSGGSTNVYY691 pKa = 10.86AFTTFQGAGVVLSGFGAYY709 pKa = 9.31PSAQALYY716 pKa = 9.44RR717 pKa = 11.84LNGGASFSASVPAAGSVTSGLEE739 pKa = 3.78VAVDD743 pKa = 3.88SNGDD747 pKa = 3.63GVFNSADD754 pKa = 3.63VFPSGASITLNISAQVKK771 pKa = 9.56
MM1 pKa = 6.91NTTPRR6 pKa = 11.84VLALMAALAAGTAAAATTNTTAGQIITNQATATFTDD42 pKa = 4.15PTTLAAATPIVSNTVQTVVLPKK64 pKa = 10.18PGFDD68 pKa = 2.85IQYY71 pKa = 10.81ADD73 pKa = 3.72GSADD77 pKa = 3.38NTTATAPAPSFDD89 pKa = 3.64KK90 pKa = 11.04TGVLPGTTVTTGYY103 pKa = 10.84VVVNTGNVNGYY114 pKa = 8.52VVNLAADD121 pKa = 3.84STGGNAPQDD130 pKa = 3.41VKK132 pKa = 11.58YY133 pKa = 11.1YY134 pKa = 11.08LDD136 pKa = 4.05ANNDD140 pKa = 3.72GVPDD144 pKa = 3.49SSTPVTSVTLNADD157 pKa = 3.86NPNTPADD164 pKa = 3.75EE165 pKa = 5.11GIVRR169 pKa = 11.84ILQVITVASTAGTGAQYY186 pKa = 10.78SASPAGTAPSGSVSATDD203 pKa = 3.29TSTGITTAYY212 pKa = 9.22PYY214 pKa = 11.04AALTEE219 pKa = 4.36AQANPASTNGDD230 pKa = 3.41LQYY233 pKa = 11.15TRR235 pKa = 11.84VTVFTPTVTNAPADD249 pKa = 4.15GDD251 pKa = 4.07PATAGQQTPTTTVVVPPSASTVIDD275 pKa = 3.89PNNPTSAPGTPSSPSDD291 pKa = 3.42PTQPGYY297 pKa = 9.62TDD299 pKa = 3.81PPVAGSTGAVAVTVVGNVQTAYY321 pKa = 10.12PPADD325 pKa = 3.39TNAAADD331 pKa = 3.56AVNFKK336 pKa = 9.38NTVTTPAGSPADD348 pKa = 3.94TVNLFPVNPALNVGDD363 pKa = 4.23AGYY366 pKa = 9.83GAPYY370 pKa = 8.48GTNNGDD376 pKa = 3.84GSFTLPDD383 pKa = 3.4GTVVRR388 pKa = 11.84FLNADD393 pKa = 3.56GSAPTLVTSPVDD405 pKa = 3.38GKK407 pKa = 10.73AYY409 pKa = 9.64PVVSVPAGGATVSYY423 pKa = 8.55ITQVVYY429 pKa = 10.45PDD431 pKa = 4.22SNSLTNPTPITVVVGADD448 pKa = 3.05SGNDD452 pKa = 3.25YY453 pKa = 11.48NLIADD458 pKa = 4.39GTTTDD463 pKa = 3.78KK464 pKa = 11.16VLPPALQFGDD474 pKa = 3.93SNGVQTPPTADD485 pKa = 3.51ASAQPSEE492 pKa = 4.42TVTPGAAVSTGTPTPGMTTDD512 pKa = 3.45SSAVFPMDD520 pKa = 2.76IANPGEE526 pKa = 4.16YY527 pKa = 10.27GDD529 pKa = 4.62TYY531 pKa = 10.21TLSGSVLVPLSNGTTATVPVKK552 pKa = 10.78YY553 pKa = 10.28VDD555 pKa = 3.31ASGTEE560 pKa = 4.09LPKK563 pKa = 10.81NAAGQYY569 pKa = 6.33ITPVVDD575 pKa = 4.63ANAEE579 pKa = 4.0YY580 pKa = 10.12RR581 pKa = 11.84VYY583 pKa = 11.0AVVDD587 pKa = 3.49IPSNARR593 pKa = 11.84LTLPGSPLSVQQTVTSNYY611 pKa = 10.8SNITLQDD618 pKa = 3.46TNDD621 pKa = 4.83LIRR624 pKa = 11.84VGAIGGITVDD634 pKa = 3.48KK635 pKa = 9.49YY636 pKa = 10.47QAVGAAPSQTAAGKK650 pKa = 8.12TIKK653 pKa = 9.4TALPGEE659 pKa = 4.48TINYY663 pKa = 9.52AIVARR668 pKa = 11.84NSYY671 pKa = 10.7NDD673 pKa = 3.29AVKK676 pKa = 10.87NFVLSDD682 pKa = 3.37VSGGSTNVYY691 pKa = 10.86AFTTFQGAGVVLSGFGAYY709 pKa = 9.31PSAQALYY716 pKa = 9.44RR717 pKa = 11.84LNGGASFSASVPAAGSVTSGLEE739 pKa = 3.78VAVDD743 pKa = 3.88SNGDD747 pKa = 3.63GVFNSADD754 pKa = 3.63VFPSGASITLNISAQVKK771 pKa = 9.56
Molecular weight: 76.94 kDa
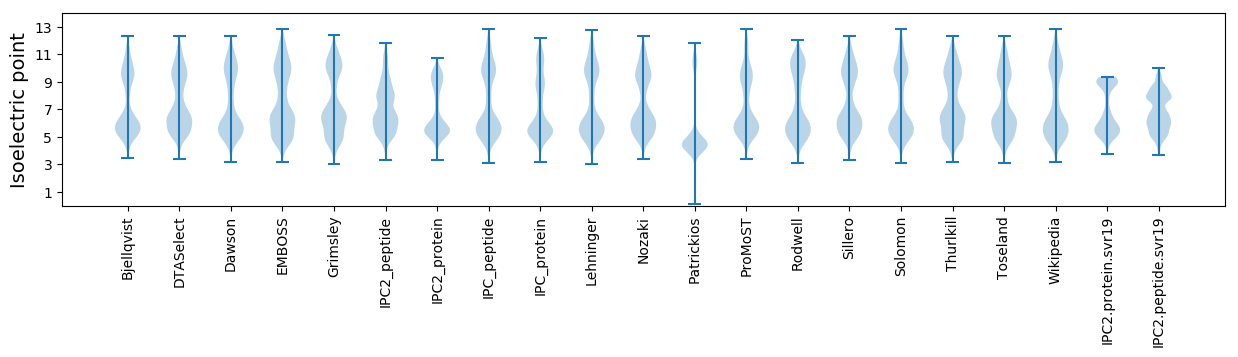
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K3V247|A0A2K3V247_9DEIO Sodium:proton antiporter OS=Deinococcus koreensis OX=2054903 GN=CVO96_03895 PE=4 SV=1
MM1 pKa = 7.13GRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84QDD8 pKa = 2.96EE9 pKa = 4.51TTVLGSSEE17 pKa = 4.3VGTDD21 pKa = 3.61LLALRR26 pKa = 11.84AQLHH30 pKa = 5.41RR31 pKa = 11.84AEE33 pKa = 4.52KK34 pKa = 8.38ATRR37 pKa = 11.84RR38 pKa = 11.84APTPPPVRR46 pKa = 11.84PQRR49 pKa = 11.84PQPVKK54 pKa = 9.28PQRR57 pKa = 11.84EE58 pKa = 4.16AEE60 pKa = 4.14LAGVLTDD67 pKa = 3.44EE68 pKa = 4.95FSLHH72 pKa = 6.29RR73 pKa = 11.84AHH75 pKa = 7.05ARR77 pKa = 11.84LRR79 pKa = 11.84AAVYY83 pKa = 10.22DD84 pKa = 4.2GAYY87 pKa = 9.68HH88 pKa = 6.99LCPHH92 pKa = 7.46AIGHH96 pKa = 6.1ARR98 pKa = 11.84AEE100 pKa = 4.37GFLEE104 pKa = 4.06HH105 pKa = 7.81DD106 pKa = 3.87ILNVLLSGRR115 pKa = 11.84VRR117 pKa = 11.84AVYY120 pKa = 10.17PDD122 pKa = 3.25DD123 pKa = 5.17HH124 pKa = 6.78RR125 pKa = 11.84WLVCGQFEE133 pKa = 4.62ACGVSLPLHH142 pKa = 5.72VVVQHH147 pKa = 5.85FRR149 pKa = 11.84SGVGGGHH156 pKa = 6.63IDD158 pKa = 2.91IVTAFVPRR166 pKa = 11.84HH167 pKa = 4.62PHH169 pKa = 5.26HH170 pKa = 6.7VISRR174 pKa = 11.84ARR176 pKa = 11.84LAVMLRR182 pKa = 11.84YY183 pKa = 9.93DD184 pKa = 3.82DD185 pKa = 3.92EE186 pKa = 4.66QIRR189 pKa = 11.84THH191 pKa = 6.06TAAPTPKK198 pKa = 9.83AGHH201 pKa = 6.81RR202 pKa = 11.84SRR204 pKa = 11.84GRR206 pKa = 11.84WKK208 pKa = 10.52KK209 pKa = 10.77SSS211 pKa = 3.16
MM1 pKa = 7.13GRR3 pKa = 11.84SRR5 pKa = 11.84RR6 pKa = 11.84QDD8 pKa = 2.96EE9 pKa = 4.51TTVLGSSEE17 pKa = 4.3VGTDD21 pKa = 3.61LLALRR26 pKa = 11.84AQLHH30 pKa = 5.41RR31 pKa = 11.84AEE33 pKa = 4.52KK34 pKa = 8.38ATRR37 pKa = 11.84RR38 pKa = 11.84APTPPPVRR46 pKa = 11.84PQRR49 pKa = 11.84PQPVKK54 pKa = 9.28PQRR57 pKa = 11.84EE58 pKa = 4.16AEE60 pKa = 4.14LAGVLTDD67 pKa = 3.44EE68 pKa = 4.95FSLHH72 pKa = 6.29RR73 pKa = 11.84AHH75 pKa = 7.05ARR77 pKa = 11.84LRR79 pKa = 11.84AAVYY83 pKa = 10.22DD84 pKa = 4.2GAYY87 pKa = 9.68HH88 pKa = 6.99LCPHH92 pKa = 7.46AIGHH96 pKa = 6.1ARR98 pKa = 11.84AEE100 pKa = 4.37GFLEE104 pKa = 4.06HH105 pKa = 7.81DD106 pKa = 3.87ILNVLLSGRR115 pKa = 11.84VRR117 pKa = 11.84AVYY120 pKa = 10.17PDD122 pKa = 3.25DD123 pKa = 5.17HH124 pKa = 6.78RR125 pKa = 11.84WLVCGQFEE133 pKa = 4.62ACGVSLPLHH142 pKa = 5.72VVVQHH147 pKa = 5.85FRR149 pKa = 11.84SGVGGGHH156 pKa = 6.63IDD158 pKa = 2.91IVTAFVPRR166 pKa = 11.84HH167 pKa = 4.62PHH169 pKa = 5.26HH170 pKa = 6.7VISRR174 pKa = 11.84ARR176 pKa = 11.84LAVMLRR182 pKa = 11.84YY183 pKa = 9.93DD184 pKa = 3.82DD185 pKa = 3.92EE186 pKa = 4.66QIRR189 pKa = 11.84THH191 pKa = 6.06TAAPTPKK198 pKa = 9.83AGHH201 pKa = 6.81RR202 pKa = 11.84SRR204 pKa = 11.84GRR206 pKa = 11.84WKK208 pKa = 10.52KK209 pKa = 10.77SSS211 pKa = 3.16
Molecular weight: 23.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1274002 |
29 |
3267 |
321.1 |
34.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.684 ± 0.053 | 0.566 ± 0.011 |
5.012 ± 0.034 | 5.317 ± 0.037 |
3.103 ± 0.024 | 9.508 ± 0.047 |
2.098 ± 0.024 | 3.398 ± 0.029 |
2.191 ± 0.03 | 11.87 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.832 ± 0.019 | 2.238 ± 0.024 |
6.027 ± 0.036 | 3.817 ± 0.025 |
7.553 ± 0.037 | 5.343 ± 0.031 |
5.941 ± 0.04 | 7.836 ± 0.029 |
1.395 ± 0.016 | 2.271 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
