
Tortoise microvirus 30
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
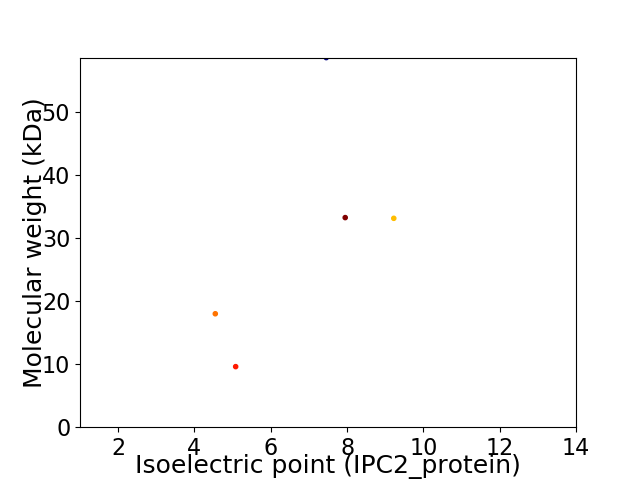
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W722|A0A4P8W722_9VIRU DNA pilot protein OS=Tortoise microvirus 30 OX=2583133 PE=4 SV=1
MM1 pKa = 7.65LIKK4 pKa = 9.33EE5 pKa = 4.62TKK7 pKa = 9.6LKK9 pKa = 10.72RR10 pKa = 11.84KK11 pKa = 9.76IITQNDD17 pKa = 3.62LQEE20 pKa = 4.34VAEE23 pKa = 4.51SLLNDD28 pKa = 3.86KK29 pKa = 9.34VTDD32 pKa = 3.59PTGYY36 pKa = 9.61IPLRR40 pKa = 11.84NRR42 pKa = 11.84IEE44 pKa = 4.0AFEE47 pKa = 4.14AAGQNLEE54 pKa = 3.86EE55 pKa = 4.34WRR57 pKa = 11.84DD58 pKa = 3.47QNFDD62 pKa = 3.66YY63 pKa = 10.92EE64 pKa = 4.43SWEE67 pKa = 3.9EE68 pKa = 3.56EE69 pKa = 4.54DD70 pKa = 4.98IIEE73 pKa = 4.24EE74 pKa = 4.05NMNLGQYY81 pKa = 10.36AIRR84 pKa = 11.84RR85 pKa = 11.84QGSDD89 pKa = 2.12IVEE92 pKa = 3.95ILNEE96 pKa = 3.69QEE98 pKa = 3.62AAYY101 pKa = 10.41FNLLRR106 pKa = 11.84QTKK109 pKa = 7.42QPRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.32LWEE118 pKa = 4.27EE119 pKa = 3.92EE120 pKa = 4.79ADD122 pKa = 5.51DD123 pKa = 5.08GDD125 pKa = 4.32PKK127 pKa = 10.91QGADD131 pKa = 3.55SKK133 pKa = 11.35KK134 pKa = 8.13EE135 pKa = 3.91HH136 pKa = 6.54QDD138 pKa = 3.23QSHH141 pKa = 6.45TDD143 pKa = 2.92SGTDD147 pKa = 3.13KK148 pKa = 11.17KK149 pKa = 9.95EE150 pKa = 3.96QKK152 pKa = 10.34DD153 pKa = 3.41
MM1 pKa = 7.65LIKK4 pKa = 9.33EE5 pKa = 4.62TKK7 pKa = 9.6LKK9 pKa = 10.72RR10 pKa = 11.84KK11 pKa = 9.76IITQNDD17 pKa = 3.62LQEE20 pKa = 4.34VAEE23 pKa = 4.51SLLNDD28 pKa = 3.86KK29 pKa = 9.34VTDD32 pKa = 3.59PTGYY36 pKa = 9.61IPLRR40 pKa = 11.84NRR42 pKa = 11.84IEE44 pKa = 4.0AFEE47 pKa = 4.14AAGQNLEE54 pKa = 3.86EE55 pKa = 4.34WRR57 pKa = 11.84DD58 pKa = 3.47QNFDD62 pKa = 3.66YY63 pKa = 10.92EE64 pKa = 4.43SWEE67 pKa = 3.9EE68 pKa = 3.56EE69 pKa = 4.54DD70 pKa = 4.98IIEE73 pKa = 4.24EE74 pKa = 4.05NMNLGQYY81 pKa = 10.36AIRR84 pKa = 11.84RR85 pKa = 11.84QGSDD89 pKa = 2.12IVEE92 pKa = 3.95ILNEE96 pKa = 3.69QEE98 pKa = 3.62AAYY101 pKa = 10.41FNLLRR106 pKa = 11.84QTKK109 pKa = 7.42QPRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84DD115 pKa = 3.32LWEE118 pKa = 4.27EE119 pKa = 3.92EE120 pKa = 4.79ADD122 pKa = 5.51DD123 pKa = 5.08GDD125 pKa = 4.32PKK127 pKa = 10.91QGADD131 pKa = 3.55SKK133 pKa = 11.35KK134 pKa = 8.13EE135 pKa = 3.91HH136 pKa = 6.54QDD138 pKa = 3.23QSHH141 pKa = 6.45TDD143 pKa = 2.92SGTDD147 pKa = 3.13KK148 pKa = 11.17KK149 pKa = 9.95EE150 pKa = 3.96QKK152 pKa = 10.34DD153 pKa = 3.41
Molecular weight: 18.0 kDa
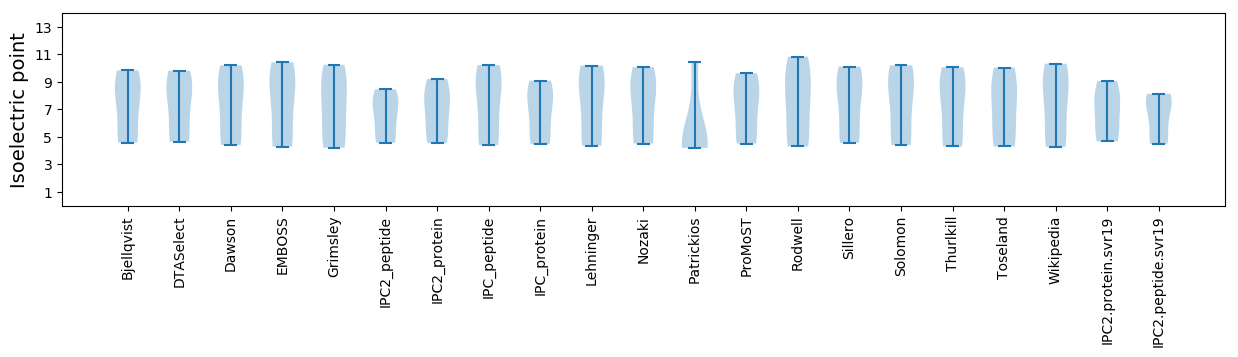
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9K2|A0A4P8W9K2_9VIRU Major capsid protein OS=Tortoise microvirus 30 OX=2583133 PE=3 SV=1
MM1 pKa = 6.29TCNRR5 pKa = 11.84KK6 pKa = 9.35FYY8 pKa = 10.1INKK11 pKa = 9.2VLVPCGKK18 pKa = 10.6CIACKK23 pKa = 10.1KK24 pKa = 9.55NKK26 pKa = 8.04SHH28 pKa = 6.17EE29 pKa = 3.95WAKK32 pKa = 10.62RR33 pKa = 11.84ISDD36 pKa = 4.7EE37 pKa = 4.34IFCSDD42 pKa = 4.11GQCSFITLTYY52 pKa = 10.33KK53 pKa = 10.6EE54 pKa = 4.12NNKK57 pKa = 9.63AQIYY61 pKa = 9.7SLSIDD66 pKa = 5.05DD67 pKa = 4.23IQKK70 pKa = 8.72FWKK73 pKa = 9.96RR74 pKa = 11.84LRR76 pKa = 11.84KK77 pKa = 9.63KK78 pKa = 10.62FPNKK82 pKa = 9.2KK83 pKa = 9.73FKK85 pKa = 10.77YY86 pKa = 8.65FCCGEE91 pKa = 4.08YY92 pKa = 10.89GEE94 pKa = 4.55QNQRR98 pKa = 11.84PHH100 pKa = 5.5YY101 pKa = 8.59HH102 pKa = 6.96AIIIGLSLKK111 pKa = 10.67DD112 pKa = 3.68LEE114 pKa = 4.63AEE116 pKa = 4.4KK117 pKa = 10.51KK118 pKa = 8.1QHH120 pKa = 6.17SNYY123 pKa = 8.71ATSKK127 pKa = 9.82VLEE130 pKa = 4.35KK131 pKa = 9.78TWPFGFNTVGIATINSIHH149 pKa = 6.04YY150 pKa = 9.26VSGYY154 pKa = 9.19ILKK157 pKa = 7.11TTWGQHH163 pKa = 4.38AVKK166 pKa = 9.92IYY168 pKa = 11.02DD169 pKa = 3.95KK170 pKa = 10.9IGLTRR175 pKa = 11.84PFLITSKK182 pKa = 10.73QLGLKK187 pKa = 8.73YY188 pKa = 10.49LKK190 pKa = 10.1EE191 pKa = 4.07NQEE194 pKa = 4.37MIRR197 pKa = 11.84NTRR200 pKa = 11.84KK201 pKa = 10.18GNPRR205 pKa = 11.84YY206 pKa = 9.61YY207 pKa = 9.73IKK209 pKa = 10.42KK210 pKa = 10.06SKK212 pKa = 10.7EE213 pKa = 3.23IDD215 pKa = 3.46SNINIIKK222 pKa = 10.07GSPKK226 pKa = 9.92IISKK230 pKa = 10.95EE231 pKa = 3.78EE232 pKa = 3.86GKK234 pKa = 10.41KK235 pKa = 10.65LKK237 pKa = 10.01TIMTKK242 pKa = 10.37NKK244 pKa = 9.13CNLMEE249 pKa = 4.14ARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84ILEE256 pKa = 4.1SNLARR261 pKa = 11.84EE262 pKa = 4.29QNEE265 pKa = 3.96LAKK268 pKa = 10.4QHH270 pKa = 6.0LLSRR274 pKa = 11.84RR275 pKa = 11.84HH276 pKa = 5.42KK277 pKa = 10.95NIGEE281 pKa = 4.4FII283 pKa = 4.07
MM1 pKa = 6.29TCNRR5 pKa = 11.84KK6 pKa = 9.35FYY8 pKa = 10.1INKK11 pKa = 9.2VLVPCGKK18 pKa = 10.6CIACKK23 pKa = 10.1KK24 pKa = 9.55NKK26 pKa = 8.04SHH28 pKa = 6.17EE29 pKa = 3.95WAKK32 pKa = 10.62RR33 pKa = 11.84ISDD36 pKa = 4.7EE37 pKa = 4.34IFCSDD42 pKa = 4.11GQCSFITLTYY52 pKa = 10.33KK53 pKa = 10.6EE54 pKa = 4.12NNKK57 pKa = 9.63AQIYY61 pKa = 9.7SLSIDD66 pKa = 5.05DD67 pKa = 4.23IQKK70 pKa = 8.72FWKK73 pKa = 9.96RR74 pKa = 11.84LRR76 pKa = 11.84KK77 pKa = 9.63KK78 pKa = 10.62FPNKK82 pKa = 9.2KK83 pKa = 9.73FKK85 pKa = 10.77YY86 pKa = 8.65FCCGEE91 pKa = 4.08YY92 pKa = 10.89GEE94 pKa = 4.55QNQRR98 pKa = 11.84PHH100 pKa = 5.5YY101 pKa = 8.59HH102 pKa = 6.96AIIIGLSLKK111 pKa = 10.67DD112 pKa = 3.68LEE114 pKa = 4.63AEE116 pKa = 4.4KK117 pKa = 10.51KK118 pKa = 8.1QHH120 pKa = 6.17SNYY123 pKa = 8.71ATSKK127 pKa = 9.82VLEE130 pKa = 4.35KK131 pKa = 9.78TWPFGFNTVGIATINSIHH149 pKa = 6.04YY150 pKa = 9.26VSGYY154 pKa = 9.19ILKK157 pKa = 7.11TTWGQHH163 pKa = 4.38AVKK166 pKa = 9.92IYY168 pKa = 11.02DD169 pKa = 3.95KK170 pKa = 10.9IGLTRR175 pKa = 11.84PFLITSKK182 pKa = 10.73QLGLKK187 pKa = 8.73YY188 pKa = 10.49LKK190 pKa = 10.1EE191 pKa = 4.07NQEE194 pKa = 4.37MIRR197 pKa = 11.84NTRR200 pKa = 11.84KK201 pKa = 10.18GNPRR205 pKa = 11.84YY206 pKa = 9.61YY207 pKa = 9.73IKK209 pKa = 10.42KK210 pKa = 10.06SKK212 pKa = 10.7EE213 pKa = 3.23IDD215 pKa = 3.46SNINIIKK222 pKa = 10.07GSPKK226 pKa = 9.92IISKK230 pKa = 10.95EE231 pKa = 3.78EE232 pKa = 3.86GKK234 pKa = 10.41KK235 pKa = 10.65LKK237 pKa = 10.01TIMTKK242 pKa = 10.37NKK244 pKa = 9.13CNLMEE249 pKa = 4.14ARR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84ILEE256 pKa = 4.1SNLARR261 pKa = 11.84EE262 pKa = 4.29QNEE265 pKa = 3.96LAKK268 pKa = 10.4QHH270 pKa = 6.0LLSRR274 pKa = 11.84RR275 pKa = 11.84HH276 pKa = 5.42KK277 pKa = 10.95NIGEE281 pKa = 4.4FII283 pKa = 4.07
Molecular weight: 33.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1320 |
83 |
513 |
264.0 |
30.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.515 ± 0.928 | 0.909 ± 0.586 |
5.455 ± 0.996 | 7.045 ± 1.492 |
3.864 ± 0.401 | 5.152 ± 0.165 |
2.273 ± 0.351 | 8.788 ± 0.572 |
7.955 ± 1.934 | 7.955 ± 0.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.879 ± 0.512 | 6.97 ± 0.287 |
3.712 ± 1.233 | 5.682 ± 0.732 |
5.833 ± 0.359 | 4.924 ± 0.56 |
6.212 ± 0.55 | 2.803 ± 0.566 |
1.667 ± 0.252 | 3.409 ± 0.318 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
