
Hubei tombus-like virus 32
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
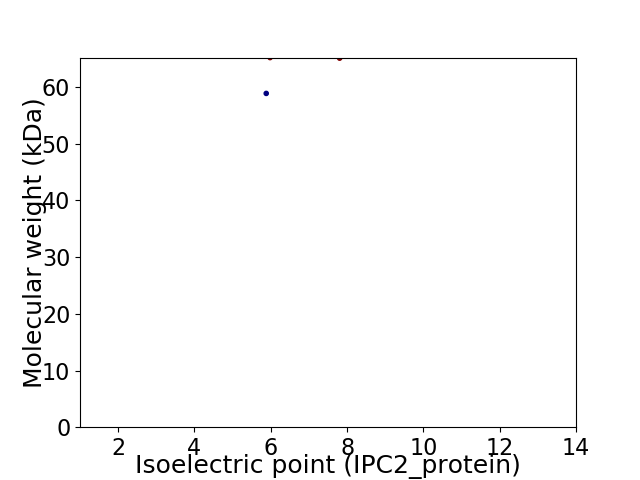
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGJ2|A0A1L3KGJ2_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 32 OX=1923280 PE=4 SV=1
MM1 pKa = 7.56DD2 pKa = 5.27HH3 pKa = 6.58KK4 pKa = 10.27HH5 pKa = 6.27LRR7 pKa = 11.84DD8 pKa = 3.55SSPVVSDD15 pKa = 4.68DD16 pKa = 4.8DD17 pKa = 4.17MEE19 pKa = 5.04VDD21 pKa = 3.85EE22 pKa = 4.87NQPGYY27 pKa = 10.24EE28 pKa = 4.32LGIAAPSNGKK38 pKa = 9.32SCLPSPVGVPMEE50 pKa = 4.24VDD52 pKa = 3.25TFIDD56 pKa = 3.95DD57 pKa = 4.45VEE59 pKa = 4.37EE60 pKa = 4.11CHH62 pKa = 7.09IGVPALLTDD71 pKa = 3.77VADD74 pKa = 4.25TLASSLRR81 pKa = 11.84LVDD84 pKa = 4.48IIIDD88 pKa = 3.62EE89 pKa = 4.44SVRR92 pKa = 11.84RR93 pKa = 11.84AVNKK97 pKa = 9.61WFDD100 pKa = 3.8NNGFIYY106 pKa = 9.41NQYY109 pKa = 9.29WDD111 pKa = 3.56EE112 pKa = 4.27DD113 pKa = 4.16ANFRR117 pKa = 11.84VDD119 pKa = 5.4DD120 pKa = 5.07DD121 pKa = 4.28EE122 pKa = 5.12LLPLARR128 pKa = 11.84MDD130 pKa = 3.65NMMSHH135 pKa = 6.35TFIPPPIVLRR145 pKa = 11.84PRR147 pKa = 11.84GARR150 pKa = 11.84YY151 pKa = 7.8TYY153 pKa = 10.24DD154 pKa = 3.32NGRR157 pKa = 11.84HH158 pKa = 3.72RR159 pKa = 11.84HH160 pKa = 4.86ARR162 pKa = 11.84LLIMSMCNPDD172 pKa = 3.36RR173 pKa = 11.84VLVRR177 pKa = 11.84SQEE180 pKa = 4.09YY181 pKa = 10.07IIEE184 pKa = 4.46KK185 pKa = 10.57NPGPPKK191 pKa = 9.87PRR193 pKa = 11.84QAAPKK198 pKa = 9.18RR199 pKa = 11.84DD200 pKa = 3.22RR201 pKa = 11.84AGPRR205 pKa = 11.84KK206 pKa = 7.69TDD208 pKa = 3.08ANRR211 pKa = 11.84GHH213 pKa = 6.6IAEE216 pKa = 4.8DD217 pKa = 3.27RR218 pKa = 11.84VVLAFRR224 pKa = 11.84LTQVYY229 pKa = 9.31VRR231 pKa = 11.84GRR233 pKa = 11.84DD234 pKa = 3.4TIYY237 pKa = 10.66PDD239 pKa = 2.69IASRR243 pKa = 11.84DD244 pKa = 3.42RR245 pKa = 11.84FNNWVRR251 pKa = 11.84ANQAQIDD258 pKa = 3.95PAEE261 pKa = 4.16MFVCKK266 pKa = 10.23ICGEE270 pKa = 4.11ASLLLCDD277 pKa = 3.52HH278 pKa = 8.06AIRR281 pKa = 11.84EE282 pKa = 4.42AVVPLACPPPVQAPTYY298 pKa = 9.43RR299 pKa = 11.84YY300 pKa = 10.1LGLRR304 pKa = 11.84RR305 pKa = 11.84LWYY308 pKa = 10.44HH309 pKa = 5.21NSKK312 pKa = 10.84FNFGEE317 pKa = 3.87QNARR321 pKa = 11.84DD322 pKa = 3.71VRR324 pKa = 11.84HH325 pKa = 6.35FDD327 pKa = 3.03NSQISDD333 pKa = 3.52VEE335 pKa = 4.47IIPEE339 pKa = 4.34CYY341 pKa = 10.43NYY343 pKa = 8.66ITSNMQTSYY352 pKa = 11.53DD353 pKa = 3.72VAGKK357 pKa = 10.09DD358 pKa = 3.53SRR360 pKa = 11.84EE361 pKa = 3.82LRR363 pKa = 11.84LSHH366 pKa = 6.05CHH368 pKa = 6.12RR369 pKa = 11.84LAGRR373 pKa = 11.84WAEE376 pKa = 3.87LAKK379 pKa = 11.11VKK381 pKa = 10.57DD382 pKa = 3.93RR383 pKa = 11.84LMEE386 pKa = 4.17DD387 pKa = 2.78TWFKK391 pKa = 11.33NRR393 pKa = 11.84LLFTAQRR400 pKa = 11.84ACDD403 pKa = 3.73QVEE406 pKa = 4.05TTMLYY411 pKa = 10.78KK412 pKa = 10.1FTDD415 pKa = 3.41PTTQNFGLARR425 pKa = 11.84QLQEE429 pKa = 3.91FVLFTTGFSALVVVLLVCGVTALEE453 pKa = 4.24FLLLLLQNIALGLSPWGLISLRR475 pKa = 11.84GKK477 pKa = 9.89IAMLACRR484 pKa = 11.84IYY486 pKa = 10.75LSKK489 pKa = 10.56LAHH492 pKa = 6.62DD493 pKa = 4.2TQSVTVRR500 pKa = 11.84VFLLCFTYY508 pKa = 8.76LTCLVDD514 pKa = 4.27ARR516 pKa = 11.84AA517 pKa = 3.79
MM1 pKa = 7.56DD2 pKa = 5.27HH3 pKa = 6.58KK4 pKa = 10.27HH5 pKa = 6.27LRR7 pKa = 11.84DD8 pKa = 3.55SSPVVSDD15 pKa = 4.68DD16 pKa = 4.8DD17 pKa = 4.17MEE19 pKa = 5.04VDD21 pKa = 3.85EE22 pKa = 4.87NQPGYY27 pKa = 10.24EE28 pKa = 4.32LGIAAPSNGKK38 pKa = 9.32SCLPSPVGVPMEE50 pKa = 4.24VDD52 pKa = 3.25TFIDD56 pKa = 3.95DD57 pKa = 4.45VEE59 pKa = 4.37EE60 pKa = 4.11CHH62 pKa = 7.09IGVPALLTDD71 pKa = 3.77VADD74 pKa = 4.25TLASSLRR81 pKa = 11.84LVDD84 pKa = 4.48IIIDD88 pKa = 3.62EE89 pKa = 4.44SVRR92 pKa = 11.84RR93 pKa = 11.84AVNKK97 pKa = 9.61WFDD100 pKa = 3.8NNGFIYY106 pKa = 9.41NQYY109 pKa = 9.29WDD111 pKa = 3.56EE112 pKa = 4.27DD113 pKa = 4.16ANFRR117 pKa = 11.84VDD119 pKa = 5.4DD120 pKa = 5.07DD121 pKa = 4.28EE122 pKa = 5.12LLPLARR128 pKa = 11.84MDD130 pKa = 3.65NMMSHH135 pKa = 6.35TFIPPPIVLRR145 pKa = 11.84PRR147 pKa = 11.84GARR150 pKa = 11.84YY151 pKa = 7.8TYY153 pKa = 10.24DD154 pKa = 3.32NGRR157 pKa = 11.84HH158 pKa = 3.72RR159 pKa = 11.84HH160 pKa = 4.86ARR162 pKa = 11.84LLIMSMCNPDD172 pKa = 3.36RR173 pKa = 11.84VLVRR177 pKa = 11.84SQEE180 pKa = 4.09YY181 pKa = 10.07IIEE184 pKa = 4.46KK185 pKa = 10.57NPGPPKK191 pKa = 9.87PRR193 pKa = 11.84QAAPKK198 pKa = 9.18RR199 pKa = 11.84DD200 pKa = 3.22RR201 pKa = 11.84AGPRR205 pKa = 11.84KK206 pKa = 7.69TDD208 pKa = 3.08ANRR211 pKa = 11.84GHH213 pKa = 6.6IAEE216 pKa = 4.8DD217 pKa = 3.27RR218 pKa = 11.84VVLAFRR224 pKa = 11.84LTQVYY229 pKa = 9.31VRR231 pKa = 11.84GRR233 pKa = 11.84DD234 pKa = 3.4TIYY237 pKa = 10.66PDD239 pKa = 2.69IASRR243 pKa = 11.84DD244 pKa = 3.42RR245 pKa = 11.84FNNWVRR251 pKa = 11.84ANQAQIDD258 pKa = 3.95PAEE261 pKa = 4.16MFVCKK266 pKa = 10.23ICGEE270 pKa = 4.11ASLLLCDD277 pKa = 3.52HH278 pKa = 8.06AIRR281 pKa = 11.84EE282 pKa = 4.42AVVPLACPPPVQAPTYY298 pKa = 9.43RR299 pKa = 11.84YY300 pKa = 10.1LGLRR304 pKa = 11.84RR305 pKa = 11.84LWYY308 pKa = 10.44HH309 pKa = 5.21NSKK312 pKa = 10.84FNFGEE317 pKa = 3.87QNARR321 pKa = 11.84DD322 pKa = 3.71VRR324 pKa = 11.84HH325 pKa = 6.35FDD327 pKa = 3.03NSQISDD333 pKa = 3.52VEE335 pKa = 4.47IIPEE339 pKa = 4.34CYY341 pKa = 10.43NYY343 pKa = 8.66ITSNMQTSYY352 pKa = 11.53DD353 pKa = 3.72VAGKK357 pKa = 10.09DD358 pKa = 3.53SRR360 pKa = 11.84EE361 pKa = 3.82LRR363 pKa = 11.84LSHH366 pKa = 6.05CHH368 pKa = 6.12RR369 pKa = 11.84LAGRR373 pKa = 11.84WAEE376 pKa = 3.87LAKK379 pKa = 11.11VKK381 pKa = 10.57DD382 pKa = 3.93RR383 pKa = 11.84LMEE386 pKa = 4.17DD387 pKa = 2.78TWFKK391 pKa = 11.33NRR393 pKa = 11.84LLFTAQRR400 pKa = 11.84ACDD403 pKa = 3.73QVEE406 pKa = 4.05TTMLYY411 pKa = 10.78KK412 pKa = 10.1FTDD415 pKa = 3.41PTTQNFGLARR425 pKa = 11.84QLQEE429 pKa = 3.91FVLFTTGFSALVVVLLVCGVTALEE453 pKa = 4.24FLLLLLQNIALGLSPWGLISLRR475 pKa = 11.84GKK477 pKa = 9.89IAMLACRR484 pKa = 11.84IYY486 pKa = 10.75LSKK489 pKa = 10.56LAHH492 pKa = 6.62DD493 pKa = 4.2TQSVTVRR500 pKa = 11.84VFLLCFTYY508 pKa = 8.76LTCLVDD514 pKa = 4.27ARR516 pKa = 11.84AA517 pKa = 3.79
Molecular weight: 58.87 kDa
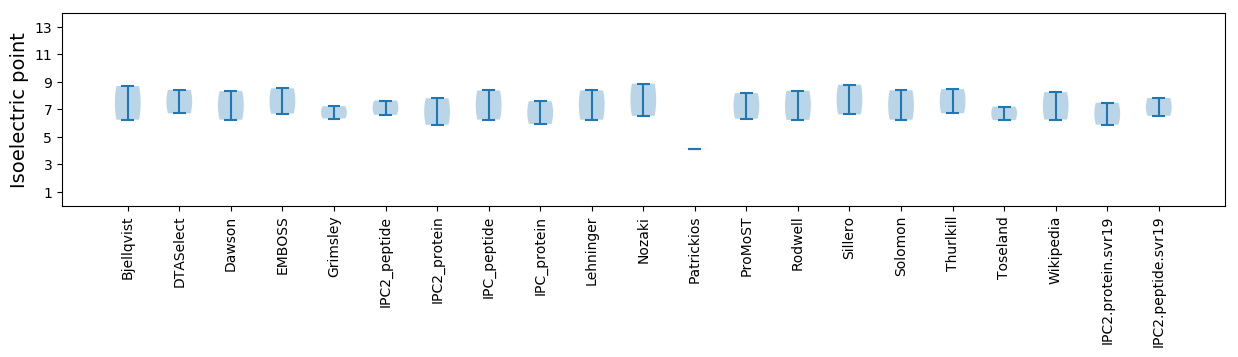
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGJ2|A0A1L3KGJ2_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 32 OX=1923280 PE=4 SV=1
MM1 pKa = 7.22QNIPVEE7 pKa = 4.19TSARR11 pKa = 11.84HH12 pKa = 5.13SKK14 pKa = 10.65CNSEE18 pKa = 4.12SFLTVFHH25 pKa = 6.78LLDD28 pKa = 3.86VPSGRR33 pKa = 11.84TGLTAGSPDD42 pKa = 4.42QITLPLLEE50 pKa = 4.66SQTTTVMSNCYY61 pKa = 9.81HH62 pKa = 5.84NQFDD66 pKa = 4.46SFTNRR71 pKa = 11.84YY72 pKa = 8.47LKK74 pKa = 9.23KK75 pKa = 9.31TPILVPRR82 pKa = 11.84NLNYY86 pKa = 10.88GLILRR91 pKa = 11.84IVKK94 pKa = 10.13HH95 pKa = 5.93LVSQVKK101 pKa = 9.01QFYY104 pKa = 10.42KK105 pKa = 10.82PEE107 pKa = 3.93FSFNKK112 pKa = 9.75YY113 pKa = 8.42VQSKK117 pKa = 9.41PGGSRR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84FLRR127 pKa = 11.84AYY129 pKa = 9.81SQLLKK134 pKa = 10.09GTNEE138 pKa = 3.79LPKK141 pKa = 9.98ISKK144 pKa = 8.2ITAFVKK150 pKa = 10.09NEE152 pKa = 3.85RR153 pKa = 11.84YY154 pKa = 9.52FDD156 pKa = 3.7EE157 pKa = 4.74SKK159 pKa = 10.35SPRR162 pKa = 11.84MIMGRR167 pKa = 11.84DD168 pKa = 3.27PRR170 pKa = 11.84FNILYY175 pKa = 9.74ARR177 pKa = 11.84FIARR181 pKa = 11.84IEE183 pKa = 3.96DD184 pKa = 3.37AFFQLEE190 pKa = 4.19QVANACDD197 pKa = 3.78FHH199 pKa = 9.08KK200 pKa = 10.79CGQKK204 pKa = 10.2FKK206 pKa = 11.47NLIGKK211 pKa = 8.52SKK213 pKa = 11.27SMFEE217 pKa = 4.2NDD219 pKa = 2.83MSKK222 pKa = 11.12YY223 pKa = 9.36EE224 pKa = 3.79ASQRR228 pKa = 11.84EE229 pKa = 3.88FALAIEE235 pKa = 4.19FLVYY239 pKa = 10.85AEE241 pKa = 4.41TVGEE245 pKa = 4.33SEE247 pKa = 4.55IDD249 pKa = 3.54DD250 pKa = 4.58LATLFAVKK258 pKa = 9.16MVKK261 pKa = 9.96SGHH264 pKa = 5.31TGEE267 pKa = 4.17GLKK270 pKa = 10.43FYY272 pKa = 10.49FLHH275 pKa = 6.56CRR277 pKa = 11.84GSGDD281 pKa = 3.39MDD283 pKa = 3.5TGLGNGVLNYY293 pKa = 8.39ITTMYY298 pKa = 10.46FKK300 pKa = 10.54IINFCPLNTDD310 pKa = 2.77CHH312 pKa = 6.56MDD314 pKa = 3.52QKK316 pKa = 11.03CCQFDD321 pKa = 3.7AFVLKK326 pKa = 10.87GDD328 pKa = 4.13DD329 pKa = 3.54SYY331 pKa = 11.79GTVPEE336 pKa = 4.37GVDD339 pKa = 3.71FNMVKK344 pKa = 8.61NTYY347 pKa = 7.87EE348 pKa = 3.82WFGFEE353 pKa = 4.24AKK355 pKa = 10.48LIHH358 pKa = 6.83RR359 pKa = 11.84PDD361 pKa = 3.16GRR363 pKa = 11.84LTEE366 pKa = 4.29FCSGNFIRR374 pKa = 11.84VAGGDD379 pKa = 3.56YY380 pKa = 10.85YY381 pKa = 11.48YY382 pKa = 11.19VQNLRR387 pKa = 11.84KK388 pKa = 9.58LVTSLTTCLNPDD400 pKa = 4.34VIRR403 pKa = 11.84EE404 pKa = 4.14GWVAHH409 pKa = 5.92YY410 pKa = 9.93YY411 pKa = 10.45RR412 pKa = 11.84SLGDD416 pKa = 3.57MYY418 pKa = 11.58AVLYY422 pKa = 10.76KK423 pKa = 10.66NIPFYY428 pKa = 11.26EE429 pKa = 5.05DD430 pKa = 2.57IAGFLQTASSKK441 pKa = 10.81LRR443 pKa = 11.84INVNLVQEE451 pKa = 4.68SYY453 pKa = 9.59GHH455 pKa = 6.98AMAFSHH461 pKa = 7.04FNRR464 pKa = 11.84NVEE467 pKa = 4.38TIDD470 pKa = 3.55ARR472 pKa = 11.84SEE474 pKa = 4.26TILDD478 pKa = 3.47ISDD481 pKa = 3.79CSGLTFPEE489 pKa = 3.99LDD491 pKa = 3.17ALRR494 pKa = 11.84TYY496 pKa = 8.01FTSSKK501 pKa = 10.15ICLPPQYY508 pKa = 10.82NRR510 pKa = 11.84RR511 pKa = 11.84CNIRR515 pKa = 11.84ATKK518 pKa = 9.78RR519 pKa = 11.84AWVDD523 pKa = 3.14LDD525 pKa = 3.57EE526 pKa = 6.52DD527 pKa = 3.4IVYY530 pKa = 10.65NINKK534 pKa = 9.92SSLTKK539 pKa = 10.16EE540 pKa = 3.72MKK542 pKa = 10.44GFRR545 pKa = 11.84KK546 pKa = 8.01TLLRR550 pKa = 11.84CLRR553 pKa = 11.84EE554 pKa = 3.86PLEE557 pKa = 4.22TLGQLAQSGTVSS569 pKa = 2.96
MM1 pKa = 7.22QNIPVEE7 pKa = 4.19TSARR11 pKa = 11.84HH12 pKa = 5.13SKK14 pKa = 10.65CNSEE18 pKa = 4.12SFLTVFHH25 pKa = 6.78LLDD28 pKa = 3.86VPSGRR33 pKa = 11.84TGLTAGSPDD42 pKa = 4.42QITLPLLEE50 pKa = 4.66SQTTTVMSNCYY61 pKa = 9.81HH62 pKa = 5.84NQFDD66 pKa = 4.46SFTNRR71 pKa = 11.84YY72 pKa = 8.47LKK74 pKa = 9.23KK75 pKa = 9.31TPILVPRR82 pKa = 11.84NLNYY86 pKa = 10.88GLILRR91 pKa = 11.84IVKK94 pKa = 10.13HH95 pKa = 5.93LVSQVKK101 pKa = 9.01QFYY104 pKa = 10.42KK105 pKa = 10.82PEE107 pKa = 3.93FSFNKK112 pKa = 9.75YY113 pKa = 8.42VQSKK117 pKa = 9.41PGGSRR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84FLRR127 pKa = 11.84AYY129 pKa = 9.81SQLLKK134 pKa = 10.09GTNEE138 pKa = 3.79LPKK141 pKa = 9.98ISKK144 pKa = 8.2ITAFVKK150 pKa = 10.09NEE152 pKa = 3.85RR153 pKa = 11.84YY154 pKa = 9.52FDD156 pKa = 3.7EE157 pKa = 4.74SKK159 pKa = 10.35SPRR162 pKa = 11.84MIMGRR167 pKa = 11.84DD168 pKa = 3.27PRR170 pKa = 11.84FNILYY175 pKa = 9.74ARR177 pKa = 11.84FIARR181 pKa = 11.84IEE183 pKa = 3.96DD184 pKa = 3.37AFFQLEE190 pKa = 4.19QVANACDD197 pKa = 3.78FHH199 pKa = 9.08KK200 pKa = 10.79CGQKK204 pKa = 10.2FKK206 pKa = 11.47NLIGKK211 pKa = 8.52SKK213 pKa = 11.27SMFEE217 pKa = 4.2NDD219 pKa = 2.83MSKK222 pKa = 11.12YY223 pKa = 9.36EE224 pKa = 3.79ASQRR228 pKa = 11.84EE229 pKa = 3.88FALAIEE235 pKa = 4.19FLVYY239 pKa = 10.85AEE241 pKa = 4.41TVGEE245 pKa = 4.33SEE247 pKa = 4.55IDD249 pKa = 3.54DD250 pKa = 4.58LATLFAVKK258 pKa = 9.16MVKK261 pKa = 9.96SGHH264 pKa = 5.31TGEE267 pKa = 4.17GLKK270 pKa = 10.43FYY272 pKa = 10.49FLHH275 pKa = 6.56CRR277 pKa = 11.84GSGDD281 pKa = 3.39MDD283 pKa = 3.5TGLGNGVLNYY293 pKa = 8.39ITTMYY298 pKa = 10.46FKK300 pKa = 10.54IINFCPLNTDD310 pKa = 2.77CHH312 pKa = 6.56MDD314 pKa = 3.52QKK316 pKa = 11.03CCQFDD321 pKa = 3.7AFVLKK326 pKa = 10.87GDD328 pKa = 4.13DD329 pKa = 3.54SYY331 pKa = 11.79GTVPEE336 pKa = 4.37GVDD339 pKa = 3.71FNMVKK344 pKa = 8.61NTYY347 pKa = 7.87EE348 pKa = 3.82WFGFEE353 pKa = 4.24AKK355 pKa = 10.48LIHH358 pKa = 6.83RR359 pKa = 11.84PDD361 pKa = 3.16GRR363 pKa = 11.84LTEE366 pKa = 4.29FCSGNFIRR374 pKa = 11.84VAGGDD379 pKa = 3.56YY380 pKa = 10.85YY381 pKa = 11.48YY382 pKa = 11.19VQNLRR387 pKa = 11.84KK388 pKa = 9.58LVTSLTTCLNPDD400 pKa = 4.34VIRR403 pKa = 11.84EE404 pKa = 4.14GWVAHH409 pKa = 5.92YY410 pKa = 9.93YY411 pKa = 10.45RR412 pKa = 11.84SLGDD416 pKa = 3.57MYY418 pKa = 11.58AVLYY422 pKa = 10.76KK423 pKa = 10.66NIPFYY428 pKa = 11.26EE429 pKa = 5.05DD430 pKa = 2.57IAGFLQTASSKK441 pKa = 10.81LRR443 pKa = 11.84INVNLVQEE451 pKa = 4.68SYY453 pKa = 9.59GHH455 pKa = 6.98AMAFSHH461 pKa = 7.04FNRR464 pKa = 11.84NVEE467 pKa = 4.38TIDD470 pKa = 3.55ARR472 pKa = 11.84SEE474 pKa = 4.26TILDD478 pKa = 3.47ISDD481 pKa = 3.79CSGLTFPEE489 pKa = 3.99LDD491 pKa = 3.17ALRR494 pKa = 11.84TYY496 pKa = 8.01FTSSKK501 pKa = 10.15ICLPPQYY508 pKa = 10.82NRR510 pKa = 11.84RR511 pKa = 11.84CNIRR515 pKa = 11.84ATKK518 pKa = 9.78RR519 pKa = 11.84AWVDD523 pKa = 3.14LDD525 pKa = 3.57EE526 pKa = 6.52DD527 pKa = 3.4IVYY530 pKa = 10.65NINKK534 pKa = 9.92SSLTKK539 pKa = 10.16EE540 pKa = 3.72MKK542 pKa = 10.44GFRR545 pKa = 11.84KK546 pKa = 8.01TLLRR550 pKa = 11.84CLRR553 pKa = 11.84EE554 pKa = 3.86PLEE557 pKa = 4.22TLGQLAQSGTVSS569 pKa = 2.96
Molecular weight: 65.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1086 |
517 |
569 |
543.0 |
61.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.446 ± 1.068 | 2.67 ± 0.027 |
6.538 ± 1.002 | 5.064 ± 0.304 |
5.433 ± 0.986 | 5.341 ± 0.642 |
2.302 ± 0.153 | 5.157 ± 0.092 |
4.788 ± 1.219 | 10.037 ± 0.433 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.486 ± 0.02 | 5.157 ± 0.37 |
4.512 ± 0.79 | 3.499 ± 0.013 |
7.182 ± 0.956 | 6.169 ± 0.821 |
5.617 ± 0.562 | 6.63 ± 0.797 |
0.921 ± 0.312 | 4.052 ± 0.549 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
