
Human gokushovirus
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
Average proteome isoelectric point is 7.64
Get precalculated fractions of proteins
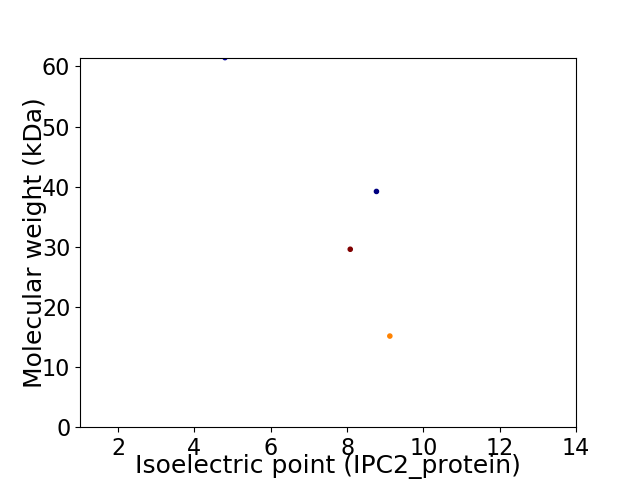
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9Q0U7|A0A1X9Q0U7_9VIRU Putative VP4 OS=Human gokushovirus OX=1986030 PE=4 SV=1
MM1 pKa = 6.93QRR3 pKa = 11.84PRR5 pKa = 11.84SKK7 pKa = 10.53FDD9 pKa = 3.45RR10 pKa = 11.84GHH12 pKa = 6.02QLLTTINEE20 pKa = 4.36GEE22 pKa = 4.28LVPIYY27 pKa = 9.68MDD29 pKa = 3.41EE30 pKa = 4.19VLPGDD35 pKa = 3.74TARR38 pKa = 11.84VQLNGLIRR46 pKa = 11.84MSTPIYY52 pKa = 9.97PIMDD56 pKa = 3.93NCYY59 pKa = 7.94MDD61 pKa = 3.49TYY63 pKa = 11.04FFFVPSRR70 pKa = 11.84LLWEE74 pKa = 4.4HH75 pKa = 6.43FEE77 pKa = 4.56NMFGEE82 pKa = 4.28NDD84 pKa = 3.07TDD86 pKa = 3.34YY87 pKa = 10.41WAEE90 pKa = 3.83KK91 pKa = 9.71TEE93 pKa = 4.34YY94 pKa = 9.2STPTCTIGGTSGLANGSIGDD114 pKa = 3.93YY115 pKa = 10.49FGLPTEE121 pKa = 4.29VNKK124 pKa = 10.55ALKK127 pKa = 10.63VNALPARR134 pKa = 11.84AYY136 pKa = 10.96AMIYY140 pKa = 10.19NEE142 pKa = 3.96WFRR145 pKa = 11.84DD146 pKa = 3.72EE147 pKa = 4.86NLEE150 pKa = 4.14APLMLGYY157 pKa = 10.7KK158 pKa = 8.98KK159 pKa = 9.19TDD161 pKa = 3.62DD162 pKa = 3.93GGTNADD168 pKa = 3.53ASEE171 pKa = 4.33EE172 pKa = 4.2TSSANAIDD180 pKa = 3.77KK181 pKa = 7.6TTNTNEE187 pKa = 3.62ATLYY191 pKa = 10.77AMKK194 pKa = 9.7PARR197 pKa = 11.84AGKK200 pKa = 8.95FHH202 pKa = 7.99DD203 pKa = 4.67YY204 pKa = 8.27FTSCLPSPMKK214 pKa = 10.71NEE216 pKa = 3.91EE217 pKa = 4.12AVTIPLGGIAPVKK230 pKa = 10.15NYY232 pKa = 10.04KK233 pKa = 10.26HH234 pKa = 6.93EE235 pKa = 4.4DD236 pKa = 3.31LKK238 pKa = 11.39DD239 pKa = 3.52PRR241 pKa = 11.84NVTLYY246 pKa = 10.35AWGAVSEE253 pKa = 4.37PGQPEE258 pKa = 3.88SGYY261 pKa = 11.62KK262 pKa = 9.55MGIQEE267 pKa = 3.95KK268 pKa = 9.83GKK270 pKa = 9.11PVKK273 pKa = 10.31IGFNDD278 pKa = 3.49NNDD281 pKa = 3.38GFLGADD287 pKa = 4.11LNNVAATTINDD298 pKa = 3.3LRR300 pKa = 11.84QAIALQHH307 pKa = 6.14IFEE310 pKa = 5.01ADD312 pKa = 3.02ARR314 pKa = 11.84NGTRR318 pKa = 11.84YY319 pKa = 9.79RR320 pKa = 11.84EE321 pKa = 4.13FLSGTWGVTSPDD333 pKa = 3.02SRR335 pKa = 11.84LQIPEE340 pKa = 4.2YY341 pKa = 10.48IGGQRR346 pKa = 11.84IAINVNQVIQTSQTDD361 pKa = 3.53AQTGQALGNTAAYY374 pKa = 10.84SLTTCSKK381 pKa = 11.1QMVDD385 pKa = 3.55YY386 pKa = 10.91AATEE390 pKa = 3.75YY391 pKa = 11.22GYY393 pKa = 10.2IIGLAVVRR401 pKa = 11.84VEE403 pKa = 4.73HH404 pKa = 7.07SYY406 pKa = 10.5QQGLGTKK413 pKa = 6.63WTRR416 pKa = 11.84GGRR419 pKa = 11.84FTYY422 pKa = 10.38YY423 pKa = 10.69DD424 pKa = 3.6PRR426 pKa = 11.84LAGLGEE432 pKa = 3.88QPVYY436 pKa = 10.71NRR438 pKa = 11.84EE439 pKa = 3.49IFAQGTEE446 pKa = 3.94KK447 pKa = 10.76DD448 pKa = 4.01DD449 pKa = 4.66EE450 pKa = 4.26IFGYY454 pKa = 7.68QEE456 pKa = 3.34CWADD460 pKa = 3.51YY461 pKa = 9.94RR462 pKa = 11.84YY463 pKa = 10.4KK464 pKa = 10.46PSYY467 pKa = 8.48VTGEE471 pKa = 3.66MRR473 pKa = 11.84SNYY476 pKa = 7.86KK477 pKa = 9.78TSLDD481 pKa = 3.19AWHH484 pKa = 6.56YY485 pKa = 11.32ADD487 pKa = 6.34DD488 pKa = 4.62YY489 pKa = 11.22DD490 pKa = 4.75TLPTLSAEE498 pKa = 4.66WIQEE502 pKa = 3.34GRR504 pKa = 11.84EE505 pKa = 4.05NIDD508 pKa = 2.82RR509 pKa = 11.84TIAVTSAVSHH519 pKa = 6.26QFLCDD524 pKa = 3.73FWFNEE529 pKa = 3.32AWFRR533 pKa = 11.84EE534 pKa = 4.03MPIYY538 pKa = 10.09SIPGIEE544 pKa = 4.29RR545 pKa = 11.84II546 pKa = 4.12
MM1 pKa = 6.93QRR3 pKa = 11.84PRR5 pKa = 11.84SKK7 pKa = 10.53FDD9 pKa = 3.45RR10 pKa = 11.84GHH12 pKa = 6.02QLLTTINEE20 pKa = 4.36GEE22 pKa = 4.28LVPIYY27 pKa = 9.68MDD29 pKa = 3.41EE30 pKa = 4.19VLPGDD35 pKa = 3.74TARR38 pKa = 11.84VQLNGLIRR46 pKa = 11.84MSTPIYY52 pKa = 9.97PIMDD56 pKa = 3.93NCYY59 pKa = 7.94MDD61 pKa = 3.49TYY63 pKa = 11.04FFFVPSRR70 pKa = 11.84LLWEE74 pKa = 4.4HH75 pKa = 6.43FEE77 pKa = 4.56NMFGEE82 pKa = 4.28NDD84 pKa = 3.07TDD86 pKa = 3.34YY87 pKa = 10.41WAEE90 pKa = 3.83KK91 pKa = 9.71TEE93 pKa = 4.34YY94 pKa = 9.2STPTCTIGGTSGLANGSIGDD114 pKa = 3.93YY115 pKa = 10.49FGLPTEE121 pKa = 4.29VNKK124 pKa = 10.55ALKK127 pKa = 10.63VNALPARR134 pKa = 11.84AYY136 pKa = 10.96AMIYY140 pKa = 10.19NEE142 pKa = 3.96WFRR145 pKa = 11.84DD146 pKa = 3.72EE147 pKa = 4.86NLEE150 pKa = 4.14APLMLGYY157 pKa = 10.7KK158 pKa = 8.98KK159 pKa = 9.19TDD161 pKa = 3.62DD162 pKa = 3.93GGTNADD168 pKa = 3.53ASEE171 pKa = 4.33EE172 pKa = 4.2TSSANAIDD180 pKa = 3.77KK181 pKa = 7.6TTNTNEE187 pKa = 3.62ATLYY191 pKa = 10.77AMKK194 pKa = 9.7PARR197 pKa = 11.84AGKK200 pKa = 8.95FHH202 pKa = 7.99DD203 pKa = 4.67YY204 pKa = 8.27FTSCLPSPMKK214 pKa = 10.71NEE216 pKa = 3.91EE217 pKa = 4.12AVTIPLGGIAPVKK230 pKa = 10.15NYY232 pKa = 10.04KK233 pKa = 10.26HH234 pKa = 6.93EE235 pKa = 4.4DD236 pKa = 3.31LKK238 pKa = 11.39DD239 pKa = 3.52PRR241 pKa = 11.84NVTLYY246 pKa = 10.35AWGAVSEE253 pKa = 4.37PGQPEE258 pKa = 3.88SGYY261 pKa = 11.62KK262 pKa = 9.55MGIQEE267 pKa = 3.95KK268 pKa = 9.83GKK270 pKa = 9.11PVKK273 pKa = 10.31IGFNDD278 pKa = 3.49NNDD281 pKa = 3.38GFLGADD287 pKa = 4.11LNNVAATTINDD298 pKa = 3.3LRR300 pKa = 11.84QAIALQHH307 pKa = 6.14IFEE310 pKa = 5.01ADD312 pKa = 3.02ARR314 pKa = 11.84NGTRR318 pKa = 11.84YY319 pKa = 9.79RR320 pKa = 11.84EE321 pKa = 4.13FLSGTWGVTSPDD333 pKa = 3.02SRR335 pKa = 11.84LQIPEE340 pKa = 4.2YY341 pKa = 10.48IGGQRR346 pKa = 11.84IAINVNQVIQTSQTDD361 pKa = 3.53AQTGQALGNTAAYY374 pKa = 10.84SLTTCSKK381 pKa = 11.1QMVDD385 pKa = 3.55YY386 pKa = 10.91AATEE390 pKa = 3.75YY391 pKa = 11.22GYY393 pKa = 10.2IIGLAVVRR401 pKa = 11.84VEE403 pKa = 4.73HH404 pKa = 7.07SYY406 pKa = 10.5QQGLGTKK413 pKa = 6.63WTRR416 pKa = 11.84GGRR419 pKa = 11.84FTYY422 pKa = 10.38YY423 pKa = 10.69DD424 pKa = 3.6PRR426 pKa = 11.84LAGLGEE432 pKa = 3.88QPVYY436 pKa = 10.71NRR438 pKa = 11.84EE439 pKa = 3.49IFAQGTEE446 pKa = 3.94KK447 pKa = 10.76DD448 pKa = 4.01DD449 pKa = 4.66EE450 pKa = 4.26IFGYY454 pKa = 7.68QEE456 pKa = 3.34CWADD460 pKa = 3.51YY461 pKa = 9.94RR462 pKa = 11.84YY463 pKa = 10.4KK464 pKa = 10.46PSYY467 pKa = 8.48VTGEE471 pKa = 3.66MRR473 pKa = 11.84SNYY476 pKa = 7.86KK477 pKa = 9.78TSLDD481 pKa = 3.19AWHH484 pKa = 6.56YY485 pKa = 11.32ADD487 pKa = 6.34DD488 pKa = 4.62YY489 pKa = 11.22DD490 pKa = 4.75TLPTLSAEE498 pKa = 4.66WIQEE502 pKa = 3.34GRR504 pKa = 11.84EE505 pKa = 4.05NIDD508 pKa = 2.82RR509 pKa = 11.84TIAVTSAVSHH519 pKa = 6.26QFLCDD524 pKa = 3.73FWFNEE529 pKa = 3.32AWFRR533 pKa = 11.84EE534 pKa = 4.03MPIYY538 pKa = 10.09SIPGIEE544 pKa = 4.29RR545 pKa = 11.84II546 pKa = 4.12
Molecular weight: 61.47 kDa
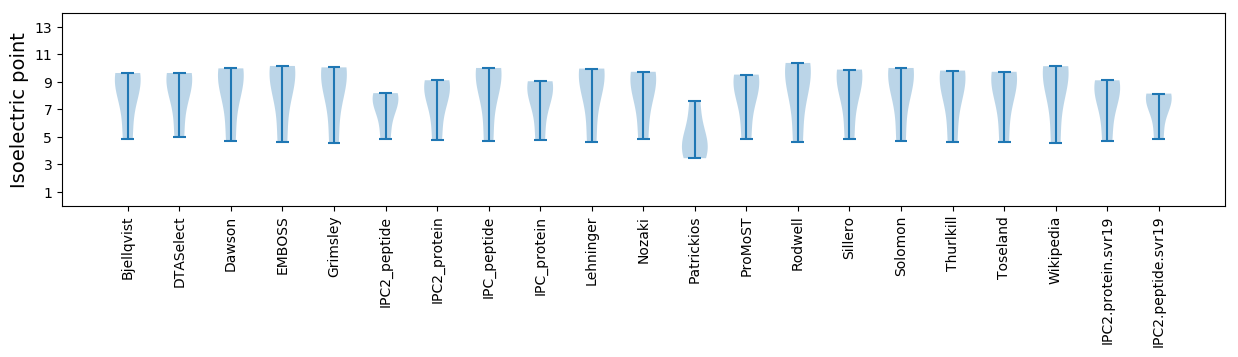
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9Q0V8|A0A1X9Q0V8_9VIRU Peptidase M15 OS=Human gokushovirus OX=1986030 PE=4 SV=1
MM1 pKa = 7.4LNEE4 pKa = 3.91YY5 pKa = 11.08SLFYY9 pKa = 10.58QGAHH13 pKa = 5.51RR14 pKa = 11.84VSAHH18 pKa = 5.57FKK20 pKa = 10.16IKK22 pKa = 10.41EE23 pKa = 3.82FAQKK27 pKa = 10.44DD28 pKa = 4.02GRR30 pKa = 11.84CDD32 pKa = 3.43KK33 pKa = 10.95ILIDD37 pKa = 3.95SEE39 pKa = 4.3LVEE42 pKa = 4.09VLEE45 pKa = 5.04DD46 pKa = 3.77VRR48 pKa = 11.84IHH50 pKa = 6.47FNAPVIITSGYY61 pKa = 7.52RR62 pKa = 11.84TPEE65 pKa = 3.74YY66 pKa = 9.43NAKK69 pKa = 10.09IGGVKK74 pKa = 10.34NSQHH78 pKa = 5.73TKK80 pKa = 8.66GTAADD85 pKa = 3.48IRR87 pKa = 11.84VKK89 pKa = 10.5GVPANKK95 pKa = 8.39VQQYY99 pKa = 10.18LKK101 pKa = 10.16HH102 pKa = 6.51KK103 pKa = 8.32YY104 pKa = 6.95WDD106 pKa = 3.47RR107 pKa = 11.84YY108 pKa = 10.54GIGSYY113 pKa = 9.84WNFTHH118 pKa = 7.7IDD120 pKa = 3.37TRR122 pKa = 11.84EE123 pKa = 3.63KK124 pKa = 9.47KK125 pKa = 10.6ARR127 pKa = 11.84WKK129 pKa = 10.5KK130 pKa = 10.27
MM1 pKa = 7.4LNEE4 pKa = 3.91YY5 pKa = 11.08SLFYY9 pKa = 10.58QGAHH13 pKa = 5.51RR14 pKa = 11.84VSAHH18 pKa = 5.57FKK20 pKa = 10.16IKK22 pKa = 10.41EE23 pKa = 3.82FAQKK27 pKa = 10.44DD28 pKa = 4.02GRR30 pKa = 11.84CDD32 pKa = 3.43KK33 pKa = 10.95ILIDD37 pKa = 3.95SEE39 pKa = 4.3LVEE42 pKa = 4.09VLEE45 pKa = 5.04DD46 pKa = 3.77VRR48 pKa = 11.84IHH50 pKa = 6.47FNAPVIITSGYY61 pKa = 7.52RR62 pKa = 11.84TPEE65 pKa = 3.74YY66 pKa = 9.43NAKK69 pKa = 10.09IGGVKK74 pKa = 10.34NSQHH78 pKa = 5.73TKK80 pKa = 8.66GTAADD85 pKa = 3.48IRR87 pKa = 11.84VKK89 pKa = 10.5GVPANKK95 pKa = 8.39VQQYY99 pKa = 10.18LKK101 pKa = 10.16HH102 pKa = 6.51KK103 pKa = 8.32YY104 pKa = 6.95WDD106 pKa = 3.47RR107 pKa = 11.84YY108 pKa = 10.54GIGSYY113 pKa = 9.84WNFTHH118 pKa = 7.7IDD120 pKa = 3.37TRR122 pKa = 11.84EE123 pKa = 3.63KK124 pKa = 9.47KK125 pKa = 10.6ARR127 pKa = 11.84WKK129 pKa = 10.5KK130 pKa = 10.27
Molecular weight: 15.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291 |
130 |
546 |
322.8 |
36.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.753 ± 1.715 | 0.93 ± 0.286 |
5.035 ± 0.621 | 6.584 ± 0.809 |
3.253 ± 0.478 | 7.746 ± 0.462 |
1.549 ± 0.446 | 5.267 ± 0.631 |
7.049 ± 1.986 | 6.429 ± 0.272 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.943 ± 0.532 | 5.964 ± 0.827 |
4.105 ± 0.511 | 4.648 ± 0.408 |
4.88 ± 0.594 | 6.274 ± 1.445 |
6.894 ± 0.899 | 4.183 ± 0.392 |
1.936 ± 0.248 | 5.577 ± 0.343 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
