
Capybara microvirus Cap1_SP_244
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.94
Get precalculated fractions of proteins
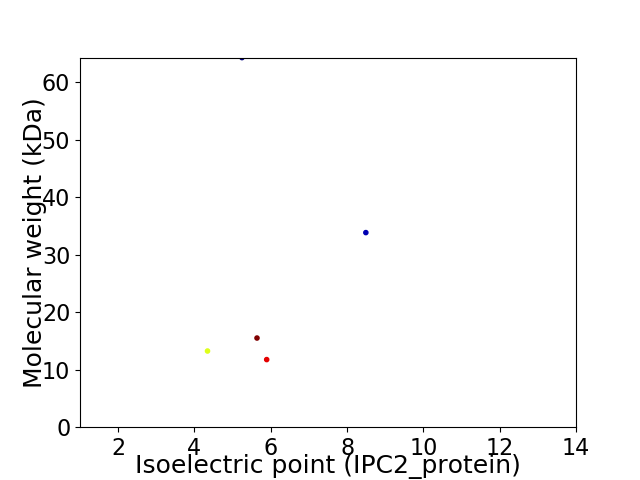
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5I1|A0A4P8W5I1_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_244 OX=2585418 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 6.07DD3 pKa = 4.35KK4 pKa = 11.63VVLPGNIDD12 pKa = 3.63SNVNYY17 pKa = 9.13DD18 pKa = 3.58ASMMPVSNLSDD29 pKa = 3.08WYY31 pKa = 10.94GEE33 pKa = 4.33SGGEE37 pKa = 4.35KK38 pKa = 10.13YY39 pKa = 10.6LQEE42 pKa = 4.16ATGSNIDD49 pKa = 4.1LAVMDD54 pKa = 4.72DD55 pKa = 4.6RR56 pKa = 11.84EE57 pKa = 4.23FEE59 pKa = 4.15LWTDD63 pKa = 3.09NTKK66 pKa = 8.84WQRR69 pKa = 11.84SVEE72 pKa = 4.02DD73 pKa = 3.48MKK75 pKa = 11.12RR76 pKa = 11.84AGINPFQALGSSPSTSVISGNQLDD100 pKa = 4.08ASGKK104 pKa = 10.35KK105 pKa = 9.89LDD107 pKa = 4.3AFLKK111 pKa = 10.76FLGTFLKK118 pKa = 11.03LII120 pKa = 4.32
MM1 pKa = 8.1DD2 pKa = 6.07DD3 pKa = 4.35KK4 pKa = 11.63VVLPGNIDD12 pKa = 3.63SNVNYY17 pKa = 9.13DD18 pKa = 3.58ASMMPVSNLSDD29 pKa = 3.08WYY31 pKa = 10.94GEE33 pKa = 4.33SGGEE37 pKa = 4.35KK38 pKa = 10.13YY39 pKa = 10.6LQEE42 pKa = 4.16ATGSNIDD49 pKa = 4.1LAVMDD54 pKa = 4.72DD55 pKa = 4.6RR56 pKa = 11.84EE57 pKa = 4.23FEE59 pKa = 4.15LWTDD63 pKa = 3.09NTKK66 pKa = 8.84WQRR69 pKa = 11.84SVEE72 pKa = 4.02DD73 pKa = 3.48MKK75 pKa = 11.12RR76 pKa = 11.84AGINPFQALGSSPSTSVISGNQLDD100 pKa = 4.08ASGKK104 pKa = 10.35KK105 pKa = 9.89LDD107 pKa = 4.3AFLKK111 pKa = 10.76FLGTFLKK118 pKa = 11.03LII120 pKa = 4.32
Molecular weight: 13.24 kDa
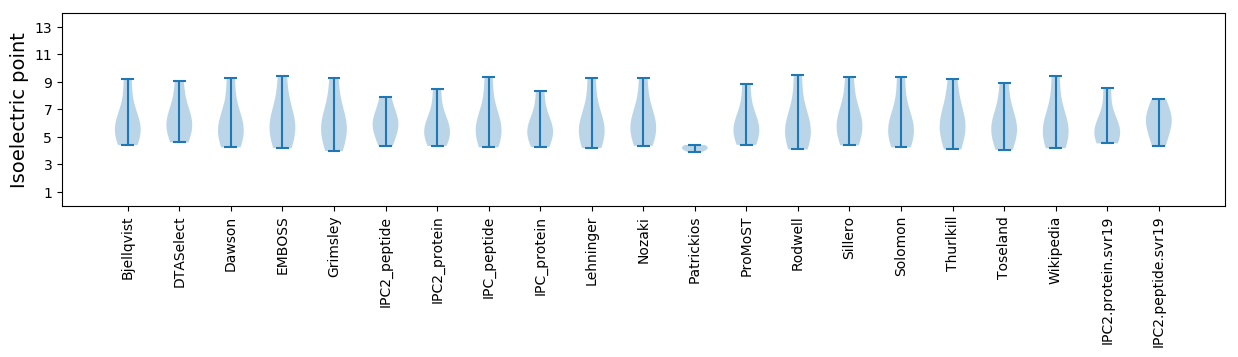
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVR5|A0A4V1FVR5_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_244 OX=2585418 PE=4 SV=1
MM1 pKa = 7.74SMNCLKK7 pKa = 10.65PKK9 pKa = 8.77MAIWNKK15 pKa = 6.99EE16 pKa = 3.8TGVFKK21 pKa = 11.08LINSDD26 pKa = 3.91DD27 pKa = 3.99LEE29 pKa = 5.0SIYY32 pKa = 11.02RR33 pKa = 11.84NSNKK37 pKa = 10.22NYY39 pKa = 8.3VPLKK43 pKa = 9.91IRR45 pKa = 11.84CGKK48 pKa = 10.32CEE50 pKa = 3.89PCNLLKK56 pKa = 11.07SQDD59 pKa = 2.78WTKK62 pKa = 11.02RR63 pKa = 11.84ILCEE67 pKa = 3.61MKK69 pKa = 10.13YY70 pKa = 10.73FKK72 pKa = 10.78YY73 pKa = 10.58SYY75 pKa = 10.24FITLTYY81 pKa = 11.02DD82 pKa = 3.07NYY84 pKa = 11.02HH85 pKa = 6.5LRR87 pKa = 11.84DD88 pKa = 3.28INTRR92 pKa = 11.84DD93 pKa = 3.02IQLFIKK99 pKa = 10.32KK100 pKa = 9.3LRR102 pKa = 11.84KK103 pKa = 10.04KK104 pKa = 9.9IDD106 pKa = 3.28RR107 pKa = 11.84KK108 pKa = 10.49LKK110 pKa = 9.42YY111 pKa = 9.21ICCGEE116 pKa = 4.16YY117 pKa = 10.61GSEE120 pKa = 4.13TKK122 pKa = 10.32RR123 pKa = 11.84PHH125 pKa = 5.82YY126 pKa = 10.74HH127 pKa = 6.59MILLSDD133 pKa = 4.09YY134 pKa = 10.93EE135 pKa = 4.35FVPPEE140 pKa = 3.47WRR142 pKa = 11.84YY143 pKa = 10.3NDD145 pKa = 3.38NTFVCSEE152 pKa = 4.79LEE154 pKa = 4.6KK155 pKa = 10.23IWQNGNVRR163 pKa = 11.84ISNDD167 pKa = 3.0VNEE170 pKa = 4.29RR171 pKa = 11.84CIKK174 pKa = 9.44YY175 pKa = 7.58TLKK178 pKa = 9.84YY179 pKa = 7.54TLKK182 pKa = 10.59NVGEE186 pKa = 4.52SKK188 pKa = 10.94VFLYY192 pKa = 10.45SKK194 pKa = 10.4KK195 pKa = 10.07IGYY198 pKa = 7.34TWFEE202 pKa = 4.16EE203 pKa = 4.08NKK205 pKa = 10.34EE206 pKa = 4.16DD207 pKa = 4.09IKK209 pKa = 11.26NYY211 pKa = 10.29CGFYY215 pKa = 10.3SKK217 pKa = 10.75NGKK220 pKa = 9.36FEE222 pKa = 4.14NPPLYY227 pKa = 10.48FIRR230 pKa = 11.84KK231 pKa = 6.94MKK233 pKa = 10.89NSDD236 pKa = 3.83DD237 pKa = 4.16FNDD240 pKa = 3.77FLWYY244 pKa = 10.13DD245 pKa = 3.71DD246 pKa = 4.08FNKK249 pKa = 10.0NRR251 pKa = 11.84LGDD254 pKa = 3.87TIIDD258 pKa = 3.94FNDD261 pKa = 2.75ILASKK266 pKa = 10.27FYY268 pKa = 10.75EE269 pKa = 4.08EE270 pKa = 4.25KK271 pKa = 10.4KK272 pKa = 9.96KK273 pKa = 11.09KK274 pKa = 9.38KK275 pKa = 10.63GKK277 pKa = 10.64GILL280 pKa = 3.5
MM1 pKa = 7.74SMNCLKK7 pKa = 10.65PKK9 pKa = 8.77MAIWNKK15 pKa = 6.99EE16 pKa = 3.8TGVFKK21 pKa = 11.08LINSDD26 pKa = 3.91DD27 pKa = 3.99LEE29 pKa = 5.0SIYY32 pKa = 11.02RR33 pKa = 11.84NSNKK37 pKa = 10.22NYY39 pKa = 8.3VPLKK43 pKa = 9.91IRR45 pKa = 11.84CGKK48 pKa = 10.32CEE50 pKa = 3.89PCNLLKK56 pKa = 11.07SQDD59 pKa = 2.78WTKK62 pKa = 11.02RR63 pKa = 11.84ILCEE67 pKa = 3.61MKK69 pKa = 10.13YY70 pKa = 10.73FKK72 pKa = 10.78YY73 pKa = 10.58SYY75 pKa = 10.24FITLTYY81 pKa = 11.02DD82 pKa = 3.07NYY84 pKa = 11.02HH85 pKa = 6.5LRR87 pKa = 11.84DD88 pKa = 3.28INTRR92 pKa = 11.84DD93 pKa = 3.02IQLFIKK99 pKa = 10.32KK100 pKa = 9.3LRR102 pKa = 11.84KK103 pKa = 10.04KK104 pKa = 9.9IDD106 pKa = 3.28RR107 pKa = 11.84KK108 pKa = 10.49LKK110 pKa = 9.42YY111 pKa = 9.21ICCGEE116 pKa = 4.16YY117 pKa = 10.61GSEE120 pKa = 4.13TKK122 pKa = 10.32RR123 pKa = 11.84PHH125 pKa = 5.82YY126 pKa = 10.74HH127 pKa = 6.59MILLSDD133 pKa = 4.09YY134 pKa = 10.93EE135 pKa = 4.35FVPPEE140 pKa = 3.47WRR142 pKa = 11.84YY143 pKa = 10.3NDD145 pKa = 3.38NTFVCSEE152 pKa = 4.79LEE154 pKa = 4.6KK155 pKa = 10.23IWQNGNVRR163 pKa = 11.84ISNDD167 pKa = 3.0VNEE170 pKa = 4.29RR171 pKa = 11.84CIKK174 pKa = 9.44YY175 pKa = 7.58TLKK178 pKa = 9.84YY179 pKa = 7.54TLKK182 pKa = 10.59NVGEE186 pKa = 4.52SKK188 pKa = 10.94VFLYY192 pKa = 10.45SKK194 pKa = 10.4KK195 pKa = 10.07IGYY198 pKa = 7.34TWFEE202 pKa = 4.16EE203 pKa = 4.08NKK205 pKa = 10.34EE206 pKa = 4.16DD207 pKa = 4.09IKK209 pKa = 11.26NYY211 pKa = 10.29CGFYY215 pKa = 10.3SKK217 pKa = 10.75NGKK220 pKa = 9.36FEE222 pKa = 4.14NPPLYY227 pKa = 10.48FIRR230 pKa = 11.84KK231 pKa = 6.94MKK233 pKa = 10.89NSDD236 pKa = 3.83DD237 pKa = 4.16FNDD240 pKa = 3.77FLWYY244 pKa = 10.13DD245 pKa = 3.71DD246 pKa = 4.08FNKK249 pKa = 10.0NRR251 pKa = 11.84LGDD254 pKa = 3.87TIIDD258 pKa = 3.94FNDD261 pKa = 2.75ILASKK266 pKa = 10.27FYY268 pKa = 10.75EE269 pKa = 4.08EE270 pKa = 4.25KK271 pKa = 10.4KK272 pKa = 9.96KK273 pKa = 11.09KK274 pKa = 9.38KK275 pKa = 10.63GKK277 pKa = 10.64GILL280 pKa = 3.5
Molecular weight: 33.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1201 |
98 |
569 |
240.2 |
27.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.83 ± 1.087 | 1.415 ± 0.716 |
7.41 ± 0.685 | 6.495 ± 0.772 |
4.913 ± 0.488 | 5.079 ± 0.631 |
1.582 ± 0.311 | 7.244 ± 0.468 |
9.992 ± 2.118 | 8.41 ± 0.414 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.165 ± 0.317 | 6.994 ± 0.682 |
3.997 ± 0.781 | 2.831 ± 0.758 |
3.164 ± 0.553 | 6.911 ± 0.887 |
5.495 ± 0.826 | 5.329 ± 0.783 |
1.166 ± 0.493 | 5.579 ± 0.863 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
