
Firmicutes bacterium CAG:646
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
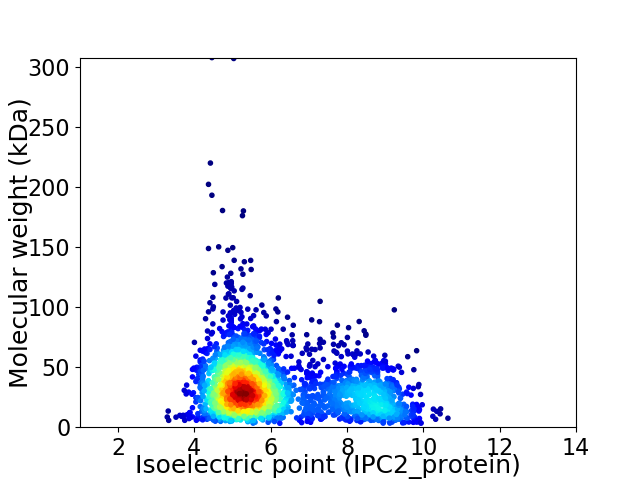
Virtual 2D-PAGE plot for 2671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5QXE0|R5QXE0_9FIRM Uncharacterized protein OS=Firmicutes bacterium CAG:646 OX=1262995 GN=BN747_02142 PE=4 SV=1
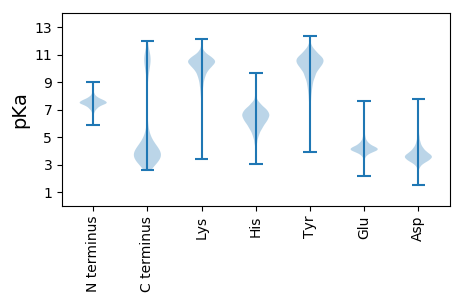
MM1 pKa = 7.55KK2 pKa = 10.13IKK4 pKa = 10.51YY5 pKa = 9.97LSVLMALGLLSAAAAGCGEE24 pKa = 4.28KK25 pKa = 10.37KK26 pKa = 10.5EE27 pKa = 4.43SFDD30 pKa = 3.78AAAYY34 pKa = 5.83TQAYY38 pKa = 9.56LDD40 pKa = 3.97ARR42 pKa = 11.84IKK44 pKa = 11.24GNTEE48 pKa = 3.58EE49 pKa = 4.11YY50 pKa = 10.76AKK52 pKa = 10.92LSGEE56 pKa = 4.2SPEE59 pKa = 4.9DD60 pKa = 3.14LAALFDD66 pKa = 4.25EE67 pKa = 5.27KK68 pKa = 11.14VQSLVTLSIGEE79 pKa = 4.14NSNEE83 pKa = 4.4PGSQLLPPQLSQDD96 pKa = 3.75YY97 pKa = 9.89AAMWKK102 pKa = 10.33DD103 pKa = 3.57VLQHH107 pKa = 5.33TKK109 pKa = 10.62YY110 pKa = 10.61KK111 pKa = 10.63AVEE114 pKa = 3.94AKK116 pKa = 10.32KK117 pKa = 10.77DD118 pKa = 3.42GDD120 pKa = 4.01SYY122 pKa = 12.03AVTIEE127 pKa = 4.14TEE129 pKa = 4.23YY130 pKa = 10.73MDD132 pKa = 6.03LNTPVNDD139 pKa = 4.12LLTEE143 pKa = 4.16KK144 pKa = 9.93LTEE147 pKa = 4.34YY148 pKa = 10.56YY149 pKa = 10.68AEE151 pKa = 4.59SPDD154 pKa = 3.58AQTNASEE161 pKa = 4.65TYY163 pKa = 9.16LQLMLEE169 pKa = 4.89SYY171 pKa = 10.7QEE173 pKa = 4.21VLQEE177 pKa = 3.89PAYY180 pKa = 9.82EE181 pKa = 4.3EE182 pKa = 4.09PAKK185 pKa = 8.85TTVTLSQDD193 pKa = 3.64DD194 pKa = 4.0KK195 pKa = 11.6KK196 pKa = 11.3VWTVSDD202 pKa = 3.99EE203 pKa = 4.23DD204 pKa = 3.94LQTVQDD210 pKa = 4.18GLFLPALYY218 pKa = 10.56SDD220 pKa = 4.74AEE222 pKa = 4.27AEE224 pKa = 4.47GEE226 pKa = 4.37GASDD230 pKa = 3.9PGSLSTEE237 pKa = 3.84ATPDD241 pKa = 3.01QTYY244 pKa = 10.79PEE246 pKa = 5.49DD247 pKa = 4.34LDD249 pKa = 3.54QTPTHH254 pKa = 6.23TLGEE258 pKa = 4.38SFILQQDD265 pKa = 3.87GQDD268 pKa = 3.18MVEE271 pKa = 4.17FSVDD275 pKa = 3.16KK276 pKa = 11.68VEE278 pKa = 4.35VTDD281 pKa = 4.12EE282 pKa = 3.94RR283 pKa = 11.84SEE285 pKa = 4.26YY286 pKa = 9.82DD287 pKa = 2.93TSNPEE292 pKa = 3.7KK293 pKa = 10.67VVVVTYY299 pKa = 8.57TYY301 pKa = 11.25KK302 pKa = 10.92NLGFADD308 pKa = 5.36PILYY312 pKa = 10.11DD313 pKa = 3.4QMSFKK318 pKa = 10.76LLEE321 pKa = 4.56GEE323 pKa = 4.82STCSPYY329 pKa = 10.95YY330 pKa = 10.27LQSLSPADD338 pKa = 3.43IATKK342 pKa = 10.69GGDD345 pKa = 3.39SVTASLAYY353 pKa = 9.9GVSADD358 pKa = 3.78CKK360 pKa = 10.02EE361 pKa = 3.96VTVYY365 pKa = 11.06VNNSQIEE372 pKa = 4.49SPFQVTVPLAA382 pKa = 3.77
MM1 pKa = 7.55KK2 pKa = 10.13IKK4 pKa = 10.51YY5 pKa = 9.97LSVLMALGLLSAAAAGCGEE24 pKa = 4.28KK25 pKa = 10.37KK26 pKa = 10.5EE27 pKa = 4.43SFDD30 pKa = 3.78AAAYY34 pKa = 5.83TQAYY38 pKa = 9.56LDD40 pKa = 3.97ARR42 pKa = 11.84IKK44 pKa = 11.24GNTEE48 pKa = 3.58EE49 pKa = 4.11YY50 pKa = 10.76AKK52 pKa = 10.92LSGEE56 pKa = 4.2SPEE59 pKa = 4.9DD60 pKa = 3.14LAALFDD66 pKa = 4.25EE67 pKa = 5.27KK68 pKa = 11.14VQSLVTLSIGEE79 pKa = 4.14NSNEE83 pKa = 4.4PGSQLLPPQLSQDD96 pKa = 3.75YY97 pKa = 9.89AAMWKK102 pKa = 10.33DD103 pKa = 3.57VLQHH107 pKa = 5.33TKK109 pKa = 10.62YY110 pKa = 10.61KK111 pKa = 10.63AVEE114 pKa = 3.94AKK116 pKa = 10.32KK117 pKa = 10.77DD118 pKa = 3.42GDD120 pKa = 4.01SYY122 pKa = 12.03AVTIEE127 pKa = 4.14TEE129 pKa = 4.23YY130 pKa = 10.73MDD132 pKa = 6.03LNTPVNDD139 pKa = 4.12LLTEE143 pKa = 4.16KK144 pKa = 9.93LTEE147 pKa = 4.34YY148 pKa = 10.56YY149 pKa = 10.68AEE151 pKa = 4.59SPDD154 pKa = 3.58AQTNASEE161 pKa = 4.65TYY163 pKa = 9.16LQLMLEE169 pKa = 4.89SYY171 pKa = 10.7QEE173 pKa = 4.21VLQEE177 pKa = 3.89PAYY180 pKa = 9.82EE181 pKa = 4.3EE182 pKa = 4.09PAKK185 pKa = 8.85TTVTLSQDD193 pKa = 3.64DD194 pKa = 4.0KK195 pKa = 11.6KK196 pKa = 11.3VWTVSDD202 pKa = 3.99EE203 pKa = 4.23DD204 pKa = 3.94LQTVQDD210 pKa = 4.18GLFLPALYY218 pKa = 10.56SDD220 pKa = 4.74AEE222 pKa = 4.27AEE224 pKa = 4.47GEE226 pKa = 4.37GASDD230 pKa = 3.9PGSLSTEE237 pKa = 3.84ATPDD241 pKa = 3.01QTYY244 pKa = 10.79PEE246 pKa = 5.49DD247 pKa = 4.34LDD249 pKa = 3.54QTPTHH254 pKa = 6.23TLGEE258 pKa = 4.38SFILQQDD265 pKa = 3.87GQDD268 pKa = 3.18MVEE271 pKa = 4.17FSVDD275 pKa = 3.16KK276 pKa = 11.68VEE278 pKa = 4.35VTDD281 pKa = 4.12EE282 pKa = 3.94RR283 pKa = 11.84SEE285 pKa = 4.26YY286 pKa = 9.82DD287 pKa = 2.93TSNPEE292 pKa = 3.7KK293 pKa = 10.67VVVVTYY299 pKa = 8.57TYY301 pKa = 11.25KK302 pKa = 10.92NLGFADD308 pKa = 5.36PILYY312 pKa = 10.11DD313 pKa = 3.4QMSFKK318 pKa = 10.76LLEE321 pKa = 4.56GEE323 pKa = 4.82STCSPYY329 pKa = 10.95YY330 pKa = 10.27LQSLSPADD338 pKa = 3.43IATKK342 pKa = 10.69GGDD345 pKa = 3.39SVTASLAYY353 pKa = 9.9GVSADD358 pKa = 3.78CKK360 pKa = 10.02EE361 pKa = 3.96VTVYY365 pKa = 11.06VNNSQIEE372 pKa = 4.49SPFQVTVPLAA382 pKa = 3.77
Molecular weight: 41.88 kDa
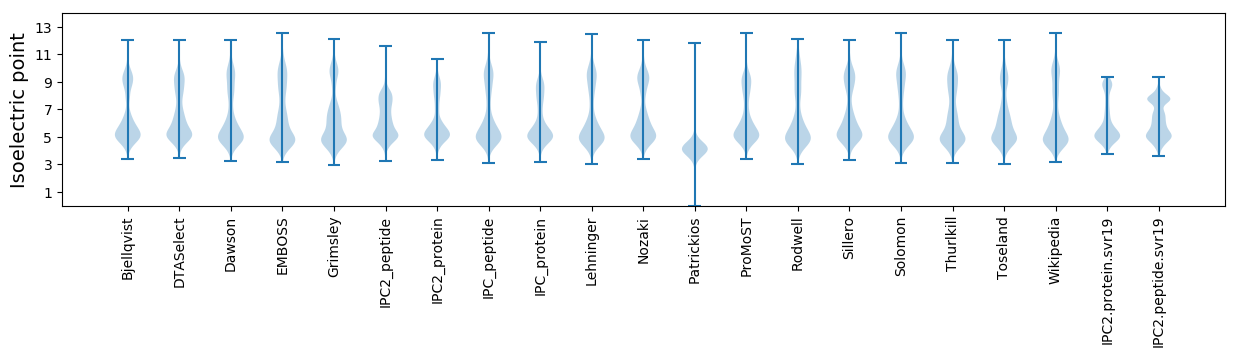
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5RMH7|R5RMH7_9FIRM Phosphoribosylaminoimidazolesuccinocarboxamide synthase OS=Firmicutes bacterium CAG:646 OX=1262995 GN=BN747_01114 PE=4 SV=1
MM1 pKa = 7.57LPTVVYY7 pKa = 10.17NYY9 pKa = 9.46RR10 pKa = 11.84TMFVQALCQIVNRR23 pKa = 11.84LCEE26 pKa = 4.21VLLHH30 pKa = 7.13LSGLDD35 pKa = 3.33LAYY38 pKa = 10.55APGFIKK44 pKa = 10.41WSPGNNTGMIVILFYY59 pKa = 11.08NFHH62 pKa = 6.87PLSDD66 pKa = 4.1KK67 pKa = 9.7FTHH70 pKa = 6.49CIIRR74 pKa = 11.84ILVSGSHH81 pKa = 6.99LAPHH85 pKa = 5.55QHH87 pKa = 6.76PFYY90 pKa = 9.62ITPIQEE96 pKa = 4.02SFIFNLLMFSQSIVTQSLNLLNIFDD121 pKa = 3.94QRR123 pKa = 11.84LLTGRR128 pKa = 11.84CQMRR132 pKa = 11.84IFPISLIQNQPLINRR147 pKa = 11.84TTVQQHH153 pKa = 5.09IRR155 pKa = 11.84TLNANISHH163 pKa = 6.21GKK165 pKa = 8.11IRR167 pKa = 11.84TYY169 pKa = 10.37RR170 pKa = 11.84VRR172 pKa = 11.84HH173 pKa = 5.83FSLEE177 pKa = 3.78HH178 pKa = 5.99HH179 pKa = 6.88FKK181 pKa = 11.31LNIIQFRR188 pKa = 11.84FFRR191 pKa = 11.84RR192 pKa = 11.84PRR194 pKa = 11.84PYY196 pKa = 11.07APFQPEE202 pKa = 3.79IIIPQEE208 pKa = 4.03YY209 pKa = 9.48IGSRR213 pKa = 11.84LHH215 pKa = 4.88MLRR218 pKa = 11.84RR219 pKa = 11.84NSRR222 pKa = 11.84EE223 pKa = 4.19HH224 pKa = 5.74FRR226 pKa = 11.84HH227 pKa = 6.46RR228 pKa = 11.84YY229 pKa = 6.92PFEE232 pKa = 4.58RR233 pKa = 11.84NRR235 pKa = 11.84CCHH238 pKa = 5.58QKK240 pKa = 9.95IFSHH244 pKa = 5.76DD245 pKa = 3.22TFRR248 pKa = 11.84RR249 pKa = 11.84LHH251 pKa = 7.09LIFQIQSMTFHH262 pKa = 7.11IRR264 pKa = 11.84RR265 pKa = 11.84HH266 pKa = 4.22FHH268 pKa = 5.66TFQMNLGNVLTPHH281 pKa = 7.49RR282 pKa = 11.84FPDD285 pKa = 3.93TGCLHH290 pKa = 6.65IPASKK295 pKa = 10.37ILIDD299 pKa = 3.84PALLSSRR306 pKa = 11.84LLHH309 pKa = 6.61IKK311 pKa = 10.34RR312 pKa = 11.84IFHH315 pKa = 6.63LHH317 pKa = 5.25HH318 pKa = 6.2QFIFSVPNYY327 pKa = 9.12RR328 pKa = 11.84SKK330 pKa = 10.66IKK332 pKa = 10.43RR333 pKa = 11.84KK334 pKa = 9.73SCVTTPVTAYY344 pKa = 9.96ILTIYY349 pKa = 10.32PNLCDD354 pKa = 5.21KK355 pKa = 10.06ITAFKK360 pKa = 9.23MKK362 pKa = 10.19HH363 pKa = 5.9IIFLFQSASIHH374 pKa = 6.21LKK376 pKa = 8.73TSSVPDD382 pKa = 3.57VIMAGFILNPTHH394 pKa = 7.01FCFISKK400 pKa = 8.16WHH402 pKa = 6.42LNGHH406 pKa = 6.27RR407 pKa = 11.84ALKK410 pKa = 10.38ISVPSLLLSTAGIIKK425 pKa = 10.21GKK427 pKa = 9.4IPRR430 pKa = 11.84SIQILPVIPHH440 pKa = 7.41KK441 pKa = 10.56LRR443 pKa = 11.84SWIILSVTLHH453 pKa = 6.42AKK455 pKa = 7.2TLRR458 pKa = 11.84FPIHH462 pKa = 6.41LNSIFVYY469 pKa = 10.59LIGLYY474 pKa = 10.41LIYY477 pKa = 9.86WVCQLNFVNHH487 pKa = 5.94KK488 pKa = 8.47MFSIIYY494 pKa = 7.83TVILSICRR502 pKa = 11.84CKK504 pKa = 9.54QHH506 pKa = 6.47EE507 pKa = 4.41YY508 pKa = 10.73CYY510 pKa = 10.38FPRR513 pKa = 11.84KK514 pKa = 9.03YY515 pKa = 10.38GFMVFPHH522 pKa = 6.88PSPTKK527 pKa = 10.01IRR529 pKa = 11.84IDD531 pKa = 3.64TGSHH535 pKa = 5.65SDD537 pKa = 3.57LFKK540 pKa = 11.0KK541 pKa = 10.51
MM1 pKa = 7.57LPTVVYY7 pKa = 10.17NYY9 pKa = 9.46RR10 pKa = 11.84TMFVQALCQIVNRR23 pKa = 11.84LCEE26 pKa = 4.21VLLHH30 pKa = 7.13LSGLDD35 pKa = 3.33LAYY38 pKa = 10.55APGFIKK44 pKa = 10.41WSPGNNTGMIVILFYY59 pKa = 11.08NFHH62 pKa = 6.87PLSDD66 pKa = 4.1KK67 pKa = 9.7FTHH70 pKa = 6.49CIIRR74 pKa = 11.84ILVSGSHH81 pKa = 6.99LAPHH85 pKa = 5.55QHH87 pKa = 6.76PFYY90 pKa = 9.62ITPIQEE96 pKa = 4.02SFIFNLLMFSQSIVTQSLNLLNIFDD121 pKa = 3.94QRR123 pKa = 11.84LLTGRR128 pKa = 11.84CQMRR132 pKa = 11.84IFPISLIQNQPLINRR147 pKa = 11.84TTVQQHH153 pKa = 5.09IRR155 pKa = 11.84TLNANISHH163 pKa = 6.21GKK165 pKa = 8.11IRR167 pKa = 11.84TYY169 pKa = 10.37RR170 pKa = 11.84VRR172 pKa = 11.84HH173 pKa = 5.83FSLEE177 pKa = 3.78HH178 pKa = 5.99HH179 pKa = 6.88FKK181 pKa = 11.31LNIIQFRR188 pKa = 11.84FFRR191 pKa = 11.84RR192 pKa = 11.84PRR194 pKa = 11.84PYY196 pKa = 11.07APFQPEE202 pKa = 3.79IIIPQEE208 pKa = 4.03YY209 pKa = 9.48IGSRR213 pKa = 11.84LHH215 pKa = 4.88MLRR218 pKa = 11.84RR219 pKa = 11.84NSRR222 pKa = 11.84EE223 pKa = 4.19HH224 pKa = 5.74FRR226 pKa = 11.84HH227 pKa = 6.46RR228 pKa = 11.84YY229 pKa = 6.92PFEE232 pKa = 4.58RR233 pKa = 11.84NRR235 pKa = 11.84CCHH238 pKa = 5.58QKK240 pKa = 9.95IFSHH244 pKa = 5.76DD245 pKa = 3.22TFRR248 pKa = 11.84RR249 pKa = 11.84LHH251 pKa = 7.09LIFQIQSMTFHH262 pKa = 7.11IRR264 pKa = 11.84RR265 pKa = 11.84HH266 pKa = 4.22FHH268 pKa = 5.66TFQMNLGNVLTPHH281 pKa = 7.49RR282 pKa = 11.84FPDD285 pKa = 3.93TGCLHH290 pKa = 6.65IPASKK295 pKa = 10.37ILIDD299 pKa = 3.84PALLSSRR306 pKa = 11.84LLHH309 pKa = 6.61IKK311 pKa = 10.34RR312 pKa = 11.84IFHH315 pKa = 6.63LHH317 pKa = 5.25HH318 pKa = 6.2QFIFSVPNYY327 pKa = 9.12RR328 pKa = 11.84SKK330 pKa = 10.66IKK332 pKa = 10.43RR333 pKa = 11.84KK334 pKa = 9.73SCVTTPVTAYY344 pKa = 9.96ILTIYY349 pKa = 10.32PNLCDD354 pKa = 5.21KK355 pKa = 10.06ITAFKK360 pKa = 9.23MKK362 pKa = 10.19HH363 pKa = 5.9IIFLFQSASIHH374 pKa = 6.21LKK376 pKa = 8.73TSSVPDD382 pKa = 3.57VIMAGFILNPTHH394 pKa = 7.01FCFISKK400 pKa = 8.16WHH402 pKa = 6.42LNGHH406 pKa = 6.27RR407 pKa = 11.84ALKK410 pKa = 10.38ISVPSLLLSTAGIIKK425 pKa = 10.21GKK427 pKa = 9.4IPRR430 pKa = 11.84SIQILPVIPHH440 pKa = 7.41KK441 pKa = 10.56LRR443 pKa = 11.84SWIILSVTLHH453 pKa = 6.42AKK455 pKa = 7.2TLRR458 pKa = 11.84FPIHH462 pKa = 6.41LNSIFVYY469 pKa = 10.59LIGLYY474 pKa = 10.41LIYY477 pKa = 9.86WVCQLNFVNHH487 pKa = 5.94KK488 pKa = 8.47MFSIIYY494 pKa = 7.83TVILSICRR502 pKa = 11.84CKK504 pKa = 9.54QHH506 pKa = 6.47EE507 pKa = 4.41YY508 pKa = 10.73CYY510 pKa = 10.38FPRR513 pKa = 11.84KK514 pKa = 9.03YY515 pKa = 10.38GFMVFPHH522 pKa = 6.88PSPTKK527 pKa = 10.01IRR529 pKa = 11.84IDD531 pKa = 3.64TGSHH535 pKa = 5.65SDD537 pKa = 3.57LFKK540 pKa = 11.0KK541 pKa = 10.51
Molecular weight: 63.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
834769 |
29 |
2951 |
312.5 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.966 ± 0.05 | 1.473 ± 0.021 |
5.194 ± 0.037 | 8.379 ± 0.064 |
4.132 ± 0.035 | 7.245 ± 0.046 |
1.801 ± 0.021 | 7.04 ± 0.045 |
6.93 ± 0.04 | 9.171 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.171 ± 0.021 | 4.023 ± 0.033 |
3.354 ± 0.025 | 3.737 ± 0.031 |
4.557 ± 0.041 | 5.505 ± 0.038 |
5.236 ± 0.043 | 6.892 ± 0.039 |
1.07 ± 0.022 | 4.121 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
