
Turnip leaf roll virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Turncurtovirus
Average proteome isoelectric point is 7.51
Get precalculated fractions of proteins
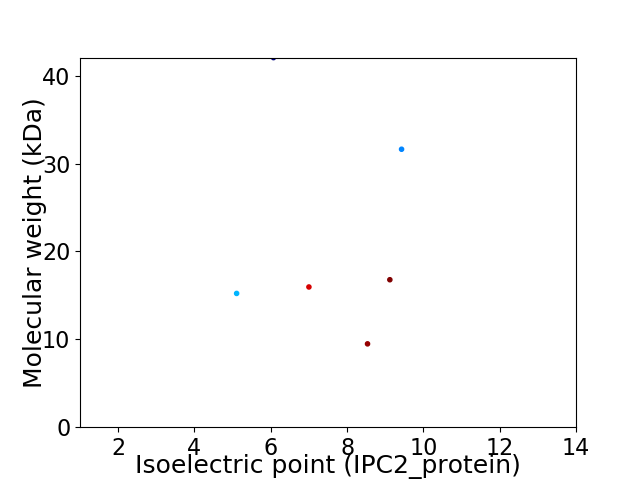
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
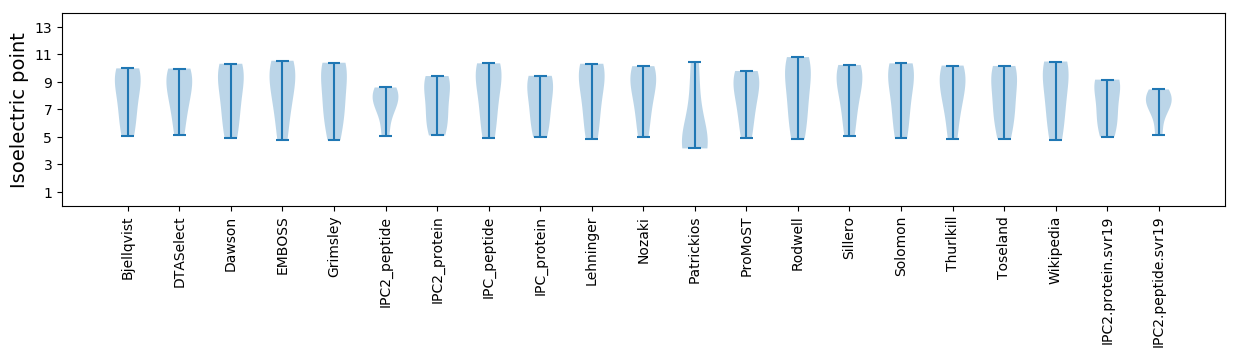
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S3JNN5|A0A0S3JNN5_9GEMI Replication-associated protein OS=Turnip leaf roll virus OX=1766828 PE=3 SV=1
MM1 pKa = 7.42IWRR4 pKa = 11.84VNKK7 pKa = 9.7DD8 pKa = 3.46DD9 pKa = 3.47KK10 pKa = 10.79RR11 pKa = 11.84VRR13 pKa = 11.84AFRR16 pKa = 11.84YY17 pKa = 8.4KK18 pKa = 10.17KK19 pKa = 9.97RR20 pKa = 11.84RR21 pKa = 11.84FYY23 pKa = 11.51SLAEE27 pKa = 3.98YY28 pKa = 10.82LLAPLATHH36 pKa = 6.85LCKK39 pKa = 10.66GRR41 pKa = 11.84IFPDD45 pKa = 3.36DD46 pKa = 4.02KK47 pKa = 11.35YY48 pKa = 11.38LVPAYY53 pKa = 9.31TDD55 pKa = 3.2EE56 pKa = 5.01AYY58 pKa = 11.07SFLVFLKK65 pKa = 10.63EE66 pKa = 3.78EE67 pKa = 4.17TRR69 pKa = 11.84TPFQILKK76 pKa = 10.45DD77 pKa = 3.52EE78 pKa = 4.46WNVEE82 pKa = 4.15FKK84 pKa = 10.63EE85 pKa = 4.18IEE87 pKa = 4.16VEE89 pKa = 4.23LSRR92 pKa = 11.84LRR94 pKa = 11.84SGPKK98 pKa = 9.86DD99 pKa = 3.33AGEE102 pKa = 3.84EE103 pKa = 4.05AGDD106 pKa = 3.99KK107 pKa = 10.46EE108 pKa = 4.76GSLSIPISVCGEE120 pKa = 3.9TSLEE124 pKa = 3.81EE125 pKa = 4.75DD126 pKa = 4.0VYY128 pKa = 11.74EE129 pKa = 4.78DD130 pKa = 3.45
MM1 pKa = 7.42IWRR4 pKa = 11.84VNKK7 pKa = 9.7DD8 pKa = 3.46DD9 pKa = 3.47KK10 pKa = 10.79RR11 pKa = 11.84VRR13 pKa = 11.84AFRR16 pKa = 11.84YY17 pKa = 8.4KK18 pKa = 10.17KK19 pKa = 9.97RR20 pKa = 11.84RR21 pKa = 11.84FYY23 pKa = 11.51SLAEE27 pKa = 3.98YY28 pKa = 10.82LLAPLATHH36 pKa = 6.85LCKK39 pKa = 10.66GRR41 pKa = 11.84IFPDD45 pKa = 3.36DD46 pKa = 4.02KK47 pKa = 11.35YY48 pKa = 11.38LVPAYY53 pKa = 9.31TDD55 pKa = 3.2EE56 pKa = 5.01AYY58 pKa = 11.07SFLVFLKK65 pKa = 10.63EE66 pKa = 3.78EE67 pKa = 4.17TRR69 pKa = 11.84TPFQILKK76 pKa = 10.45DD77 pKa = 3.52EE78 pKa = 4.46WNVEE82 pKa = 4.15FKK84 pKa = 10.63EE85 pKa = 4.18IEE87 pKa = 4.16VEE89 pKa = 4.23LSRR92 pKa = 11.84LRR94 pKa = 11.84SGPKK98 pKa = 9.86DD99 pKa = 3.33AGEE102 pKa = 3.84EE103 pKa = 4.05AGDD106 pKa = 3.99KK107 pKa = 10.46EE108 pKa = 4.76GSLSIPISVCGEE120 pKa = 3.9TSLEE124 pKa = 3.81EE125 pKa = 4.75DD126 pKa = 4.0VYY128 pKa = 11.74EE129 pKa = 4.78DD130 pKa = 3.45
Molecular weight: 15.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S3JNV6|A0A0S3JNV6_9GEMI Capsid protein OS=Turnip leaf roll virus OX=1766828 PE=3 SV=1
MM1 pKa = 7.45SGTWSLKK8 pKa = 8.4RR9 pKa = 11.84SRR11 pKa = 11.84WNSPGYY17 pKa = 10.47DD18 pKa = 3.8LAPKK22 pKa = 9.63TPAKK26 pKa = 10.23RR27 pKa = 11.84PAIRR31 pKa = 11.84RR32 pKa = 11.84ALFQSPSQYY41 pKa = 9.5VARR44 pKa = 11.84RR45 pKa = 11.84PWRR48 pKa = 11.84RR49 pKa = 11.84TFTRR53 pKa = 11.84TNKK56 pKa = 9.87GVSKK60 pKa = 10.61KK61 pKa = 10.1KK62 pKa = 10.65LKK64 pKa = 10.85ASDD67 pKa = 3.5YY68 pKa = 10.58QIYY71 pKa = 9.38EE72 pKa = 4.14DD73 pKa = 5.59RR74 pKa = 11.84VGGDD78 pKa = 3.3NGWTVTSTGDD88 pKa = 3.27CTMLNNYY95 pKa = 8.7VRR97 pKa = 11.84GIGRR101 pKa = 11.84DD102 pKa = 3.55QRR104 pKa = 11.84DD105 pKa = 3.31STLTKK110 pKa = 8.64TVHH113 pKa = 4.5MHH115 pKa = 5.93FSGVLMANDD124 pKa = 4.5AFWEE128 pKa = 4.53APNYY132 pKa = 6.13MTMYY136 pKa = 10.36SWIILDD142 pKa = 3.98NDD144 pKa = 4.09PGGSFPKK151 pKa = 10.22PADD154 pKa = 3.33IFDD157 pKa = 3.9MTDD160 pKa = 2.63KK161 pKa = 10.75DD162 pKa = 4.39FPSMYY167 pKa = 10.11EE168 pKa = 3.66VAEE171 pKa = 4.23SVKK174 pKa = 10.27PRR176 pKa = 11.84FIVKK180 pKa = 10.11RR181 pKa = 11.84KK182 pKa = 6.14TRR184 pKa = 11.84HH185 pKa = 4.59YY186 pKa = 10.76LRR188 pKa = 11.84SCGVAFGEE196 pKa = 4.26KK197 pKa = 9.6QNYY200 pKa = 7.5KK201 pKa = 10.67APTLGPIKK209 pKa = 10.41KK210 pKa = 8.76PISMVFRR217 pKa = 11.84NMWQMTEE224 pKa = 3.78WKK226 pKa = 8.6DD227 pKa = 3.55TAGGKK232 pKa = 10.06YY233 pKa = 10.11EE234 pKa = 4.17DD235 pKa = 4.21LKK237 pKa = 11.27KK238 pKa = 10.78GALLFVCMSDD248 pKa = 3.17NKK250 pKa = 10.61ASQFSFSLRR259 pKa = 11.84GKK261 pKa = 8.49WKK263 pKa = 9.22MYY265 pKa = 9.86FINRR269 pKa = 11.84DD270 pKa = 3.13RR271 pKa = 11.84FQQ273 pKa = 5.04
MM1 pKa = 7.45SGTWSLKK8 pKa = 8.4RR9 pKa = 11.84SRR11 pKa = 11.84WNSPGYY17 pKa = 10.47DD18 pKa = 3.8LAPKK22 pKa = 9.63TPAKK26 pKa = 10.23RR27 pKa = 11.84PAIRR31 pKa = 11.84RR32 pKa = 11.84ALFQSPSQYY41 pKa = 9.5VARR44 pKa = 11.84RR45 pKa = 11.84PWRR48 pKa = 11.84RR49 pKa = 11.84TFTRR53 pKa = 11.84TNKK56 pKa = 9.87GVSKK60 pKa = 10.61KK61 pKa = 10.1KK62 pKa = 10.65LKK64 pKa = 10.85ASDD67 pKa = 3.5YY68 pKa = 10.58QIYY71 pKa = 9.38EE72 pKa = 4.14DD73 pKa = 5.59RR74 pKa = 11.84VGGDD78 pKa = 3.3NGWTVTSTGDD88 pKa = 3.27CTMLNNYY95 pKa = 8.7VRR97 pKa = 11.84GIGRR101 pKa = 11.84DD102 pKa = 3.55QRR104 pKa = 11.84DD105 pKa = 3.31STLTKK110 pKa = 8.64TVHH113 pKa = 4.5MHH115 pKa = 5.93FSGVLMANDD124 pKa = 4.5AFWEE128 pKa = 4.53APNYY132 pKa = 6.13MTMYY136 pKa = 10.36SWIILDD142 pKa = 3.98NDD144 pKa = 4.09PGGSFPKK151 pKa = 10.22PADD154 pKa = 3.33IFDD157 pKa = 3.9MTDD160 pKa = 2.63KK161 pKa = 10.75DD162 pKa = 4.39FPSMYY167 pKa = 10.11EE168 pKa = 3.66VAEE171 pKa = 4.23SVKK174 pKa = 10.27PRR176 pKa = 11.84FIVKK180 pKa = 10.11RR181 pKa = 11.84KK182 pKa = 6.14TRR184 pKa = 11.84HH185 pKa = 4.59YY186 pKa = 10.76LRR188 pKa = 11.84SCGVAFGEE196 pKa = 4.26KK197 pKa = 9.6QNYY200 pKa = 7.5KK201 pKa = 10.67APTLGPIKK209 pKa = 10.41KK210 pKa = 8.76PISMVFRR217 pKa = 11.84NMWQMTEE224 pKa = 3.78WKK226 pKa = 8.6DD227 pKa = 3.55TAGGKK232 pKa = 10.06YY233 pKa = 10.11EE234 pKa = 4.17DD235 pKa = 4.21LKK237 pKa = 11.27KK238 pKa = 10.78GALLFVCMSDD248 pKa = 3.17NKK250 pKa = 10.61ASQFSFSLRR259 pKa = 11.84GKK261 pKa = 8.49WKK263 pKa = 9.22MYY265 pKa = 9.86FINRR269 pKa = 11.84DD270 pKa = 3.13RR271 pKa = 11.84FQQ273 pKa = 5.04
Molecular weight: 31.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1140 |
85 |
371 |
190.0 |
21.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.263 ± 0.962 | 1.228 ± 0.155 |
5.263 ± 0.65 | 6.053 ± 1.087 |
4.737 ± 0.503 | 5.702 ± 0.369 |
3.333 ± 0.811 | 6.404 ± 0.984 |
7.018 ± 0.736 | 7.719 ± 0.84 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.281 ± 0.885 | 5.088 ± 0.666 |
5.965 ± 0.888 | 3.86 ± 0.534 |
6.228 ± 0.825 | 7.193 ± 0.679 |
6.404 ± 0.858 | 4.561 ± 0.554 |
1.93 ± 0.383 | 3.772 ± 0.355 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
