
Roseomonas rosea
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
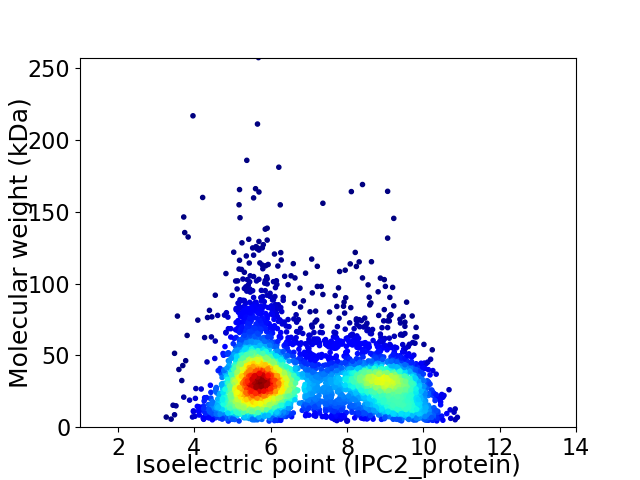
Virtual 2D-PAGE plot for 5011 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6MB58|A0A1M6MB58_9PROT Dolichol-phosphate mannosyltransferase OS=Roseomonas rosea OX=198092 GN=SAMN02745194_03415 PE=4 SV=1
MM1 pKa = 7.9PATTSPSASGTPAIDD16 pKa = 3.55GLLAGTAWAGHH27 pKa = 5.6SLTYY31 pKa = 10.53SFPGSAADD39 pKa = 4.01LSDD42 pKa = 3.59YY43 pKa = 10.68SAPNAVDD50 pKa = 3.61PSYY53 pKa = 11.45VAALSSGQQDD63 pKa = 3.81TVRR66 pKa = 11.84GMLVAFGAVANITFSEE82 pKa = 4.93AITPSGGDD90 pKa = 3.33LRR92 pKa = 11.84LYY94 pKa = 9.86WYY96 pKa = 7.65EE97 pKa = 4.35APDD100 pKa = 3.53NYY102 pKa = 9.82TARR105 pKa = 11.84TVEE108 pKa = 4.45FPSANPEE115 pKa = 4.12AGDD118 pKa = 3.85VQLGAYY124 pKa = 7.51VVPGFVDD131 pKa = 3.0QWTLGSYY138 pKa = 10.46SYY140 pKa = 9.47FTILHH145 pKa = 5.95EE146 pKa = 4.91LGHH149 pKa = 6.52ALGLKK154 pKa = 9.8HH155 pKa = 6.56PHH157 pKa = 6.93DD158 pKa = 4.68EE159 pKa = 4.6VNGFPPIPAAQDD171 pKa = 3.57SIEE174 pKa = 3.96YY175 pKa = 10.47SIMSYY180 pKa = 10.43RR181 pKa = 11.84SYY183 pKa = 11.2AGQDD187 pKa = 3.08FGGYY191 pKa = 6.37TVQSGSYY198 pKa = 9.79PSGPMLNDD206 pKa = 3.21IAALQYY212 pKa = 11.02LYY214 pKa = 10.89GANWTTNSGDD224 pKa = 3.05TTYY227 pKa = 11.21RR228 pKa = 11.84FDD230 pKa = 3.85PEE232 pKa = 3.98QAVVFQTIWDD242 pKa = 3.87GGGEE246 pKa = 4.06DD247 pKa = 4.3TYY249 pKa = 11.89DD250 pKa = 3.44LSAYY254 pKa = 8.44GTDD257 pKa = 4.13LVISLDD263 pKa = 3.42PGGWSRR269 pKa = 11.84FGGQYY274 pKa = 11.07AVLDD278 pKa = 4.08TQDD281 pKa = 2.86GRR283 pKa = 11.84EE284 pKa = 4.31AIGNVANAYY293 pKa = 9.98LYY295 pKa = 10.76QGDD298 pKa = 4.22LRR300 pKa = 11.84SLIEE304 pKa = 3.87NAIGGSGNDD313 pKa = 4.04EE314 pKa = 3.75ITGNAGANALTGGAGDD330 pKa = 3.59DD331 pKa = 3.83TLRR334 pKa = 11.84GGLGDD339 pKa = 3.69DD340 pKa = 3.82VLRR343 pKa = 11.84GGSGIDD349 pKa = 3.43LMVGGEE355 pKa = 4.24GSDD358 pKa = 3.64TYY360 pKa = 11.21HH361 pKa = 6.72VDD363 pKa = 3.28SVGDD367 pKa = 3.73RR368 pKa = 11.84VRR370 pKa = 11.84EE371 pKa = 3.9AAGEE375 pKa = 4.27GGDD378 pKa = 4.06VVWTGVDD385 pKa = 3.0WTLEE389 pKa = 3.79AGQAVEE395 pKa = 4.71VIRR398 pKa = 11.84ANAGEE403 pKa = 4.44AGLTLTGNEE412 pKa = 3.96LGNWLVSGAGADD424 pKa = 3.87LLIGGLGNDD433 pKa = 3.26SLYY436 pKa = 11.42VNSAEE441 pKa = 4.07DD442 pKa = 3.23RR443 pKa = 11.84VVEE446 pKa = 4.34LVGQGSDD453 pKa = 3.31TVFASASYY461 pKa = 9.46TLAAGQEE468 pKa = 4.14IEE470 pKa = 4.37ILRR473 pKa = 11.84ANAGATGLVLAGNDD487 pKa = 2.85IANRR491 pKa = 11.84IVGGDD496 pKa = 3.53GADD499 pKa = 3.56TLSGALGQDD508 pKa = 2.88TLMGGEE514 pKa = 4.11GADD517 pKa = 3.23VFRR520 pKa = 11.84FSSLAEE526 pKa = 4.32SPASMLRR533 pKa = 11.84DD534 pKa = 3.78YY535 pKa = 11.18ISDD538 pKa = 3.95FVSGVDD544 pKa = 5.5LINLSAIQAVAGAAVDD560 pKa = 3.51AAFTFLDD567 pKa = 3.71KK568 pKa = 11.42DD569 pKa = 3.91SFTKK573 pKa = 10.5QAGQLHH579 pKa = 5.61QIKK582 pKa = 10.24AGSNTVVEE590 pKa = 4.48GDD592 pKa = 3.62VNGDD596 pKa = 2.97GRR598 pKa = 11.84ADD600 pKa = 3.62LQILLKK606 pKa = 10.89GHH608 pKa = 7.44LDD610 pKa = 3.68LEE612 pKa = 5.34AGDD615 pKa = 4.39FLLL618 pKa = 5.79
MM1 pKa = 7.9PATTSPSASGTPAIDD16 pKa = 3.55GLLAGTAWAGHH27 pKa = 5.6SLTYY31 pKa = 10.53SFPGSAADD39 pKa = 4.01LSDD42 pKa = 3.59YY43 pKa = 10.68SAPNAVDD50 pKa = 3.61PSYY53 pKa = 11.45VAALSSGQQDD63 pKa = 3.81TVRR66 pKa = 11.84GMLVAFGAVANITFSEE82 pKa = 4.93AITPSGGDD90 pKa = 3.33LRR92 pKa = 11.84LYY94 pKa = 9.86WYY96 pKa = 7.65EE97 pKa = 4.35APDD100 pKa = 3.53NYY102 pKa = 9.82TARR105 pKa = 11.84TVEE108 pKa = 4.45FPSANPEE115 pKa = 4.12AGDD118 pKa = 3.85VQLGAYY124 pKa = 7.51VVPGFVDD131 pKa = 3.0QWTLGSYY138 pKa = 10.46SYY140 pKa = 9.47FTILHH145 pKa = 5.95EE146 pKa = 4.91LGHH149 pKa = 6.52ALGLKK154 pKa = 9.8HH155 pKa = 6.56PHH157 pKa = 6.93DD158 pKa = 4.68EE159 pKa = 4.6VNGFPPIPAAQDD171 pKa = 3.57SIEE174 pKa = 3.96YY175 pKa = 10.47SIMSYY180 pKa = 10.43RR181 pKa = 11.84SYY183 pKa = 11.2AGQDD187 pKa = 3.08FGGYY191 pKa = 6.37TVQSGSYY198 pKa = 9.79PSGPMLNDD206 pKa = 3.21IAALQYY212 pKa = 11.02LYY214 pKa = 10.89GANWTTNSGDD224 pKa = 3.05TTYY227 pKa = 11.21RR228 pKa = 11.84FDD230 pKa = 3.85PEE232 pKa = 3.98QAVVFQTIWDD242 pKa = 3.87GGGEE246 pKa = 4.06DD247 pKa = 4.3TYY249 pKa = 11.89DD250 pKa = 3.44LSAYY254 pKa = 8.44GTDD257 pKa = 4.13LVISLDD263 pKa = 3.42PGGWSRR269 pKa = 11.84FGGQYY274 pKa = 11.07AVLDD278 pKa = 4.08TQDD281 pKa = 2.86GRR283 pKa = 11.84EE284 pKa = 4.31AIGNVANAYY293 pKa = 9.98LYY295 pKa = 10.76QGDD298 pKa = 4.22LRR300 pKa = 11.84SLIEE304 pKa = 3.87NAIGGSGNDD313 pKa = 4.04EE314 pKa = 3.75ITGNAGANALTGGAGDD330 pKa = 3.59DD331 pKa = 3.83TLRR334 pKa = 11.84GGLGDD339 pKa = 3.69DD340 pKa = 3.82VLRR343 pKa = 11.84GGSGIDD349 pKa = 3.43LMVGGEE355 pKa = 4.24GSDD358 pKa = 3.64TYY360 pKa = 11.21HH361 pKa = 6.72VDD363 pKa = 3.28SVGDD367 pKa = 3.73RR368 pKa = 11.84VRR370 pKa = 11.84EE371 pKa = 3.9AAGEE375 pKa = 4.27GGDD378 pKa = 4.06VVWTGVDD385 pKa = 3.0WTLEE389 pKa = 3.79AGQAVEE395 pKa = 4.71VIRR398 pKa = 11.84ANAGEE403 pKa = 4.44AGLTLTGNEE412 pKa = 3.96LGNWLVSGAGADD424 pKa = 3.87LLIGGLGNDD433 pKa = 3.26SLYY436 pKa = 11.42VNSAEE441 pKa = 4.07DD442 pKa = 3.23RR443 pKa = 11.84VVEE446 pKa = 4.34LVGQGSDD453 pKa = 3.31TVFASASYY461 pKa = 9.46TLAAGQEE468 pKa = 4.14IEE470 pKa = 4.37ILRR473 pKa = 11.84ANAGATGLVLAGNDD487 pKa = 2.85IANRR491 pKa = 11.84IVGGDD496 pKa = 3.53GADD499 pKa = 3.56TLSGALGQDD508 pKa = 2.88TLMGGEE514 pKa = 4.11GADD517 pKa = 3.23VFRR520 pKa = 11.84FSSLAEE526 pKa = 4.32SPASMLRR533 pKa = 11.84DD534 pKa = 3.78YY535 pKa = 11.18ISDD538 pKa = 3.95FVSGVDD544 pKa = 5.5LINLSAIQAVAGAAVDD560 pKa = 3.51AAFTFLDD567 pKa = 3.71KK568 pKa = 11.42DD569 pKa = 3.91SFTKK573 pKa = 10.5QAGQLHH579 pKa = 5.61QIKK582 pKa = 10.24AGSNTVVEE590 pKa = 4.48GDD592 pKa = 3.62VNGDD596 pKa = 2.97GRR598 pKa = 11.84ADD600 pKa = 3.62LQILLKK606 pKa = 10.89GHH608 pKa = 7.44LDD610 pKa = 3.68LEE612 pKa = 5.34AGDD615 pKa = 4.39FLLL618 pKa = 5.79
Molecular weight: 63.87 kDa
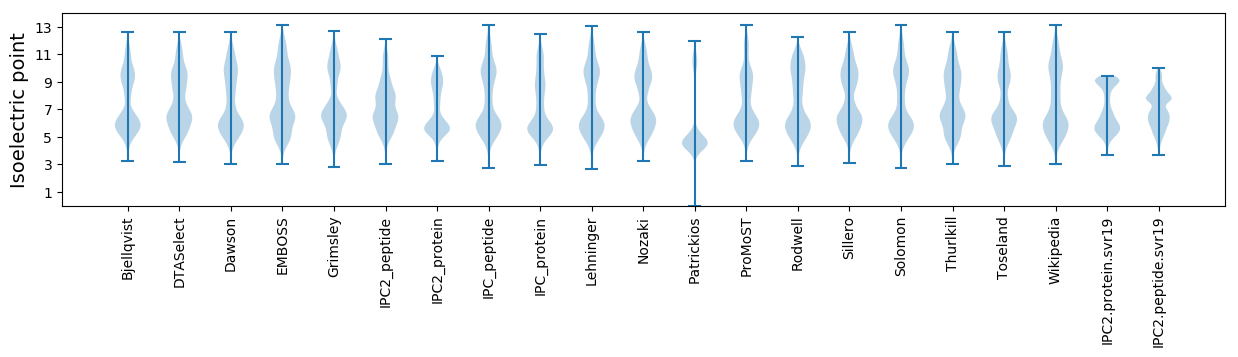
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6Q9A7|A0A1M6Q9A7_9PROT General L-amino acid transport system substrate-binding protein OS=Roseomonas rosea OX=198092 GN=SAMN02745194_04339 PE=3 SV=1
MM1 pKa = 7.34LVGRR5 pKa = 11.84IAKK8 pKa = 9.96ALIGAAIRR16 pKa = 11.84RR17 pKa = 11.84QQNRR21 pKa = 11.84ALGSIGLGGWKK32 pKa = 9.78LHH34 pKa = 6.6LFRR37 pKa = 11.84AVQAAMRR44 pKa = 11.84QGRR47 pKa = 11.84RR48 pKa = 11.84GPGQPMPRR56 pKa = 11.84PPRR59 pKa = 11.84RR60 pKa = 11.84DD61 pKa = 2.91IGG63 pKa = 3.55
MM1 pKa = 7.34LVGRR5 pKa = 11.84IAKK8 pKa = 9.96ALIGAAIRR16 pKa = 11.84RR17 pKa = 11.84QQNRR21 pKa = 11.84ALGSIGLGGWKK32 pKa = 9.78LHH34 pKa = 6.6LFRR37 pKa = 11.84AVQAAMRR44 pKa = 11.84QGRR47 pKa = 11.84RR48 pKa = 11.84GPGQPMPRR56 pKa = 11.84PPRR59 pKa = 11.84RR60 pKa = 11.84DD61 pKa = 2.91IGG63 pKa = 3.55
Molecular weight: 6.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1607065 |
39 |
2482 |
320.7 |
34.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.729 ± 0.055 | 0.768 ± 0.009 |
4.94 ± 0.024 | 5.845 ± 0.033 |
3.207 ± 0.019 | 9.496 ± 0.036 |
1.999 ± 0.017 | 4.026 ± 0.023 |
1.907 ± 0.023 | 10.926 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.439 ± 0.016 | 2.047 ± 0.016 |
6.412 ± 0.027 | 2.815 ± 0.017 |
8.556 ± 0.036 | 4.592 ± 0.021 |
4.896 ± 0.021 | 7.243 ± 0.026 |
1.42 ± 0.015 | 1.738 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
