
Mycobacterium saskatchewanense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium;
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
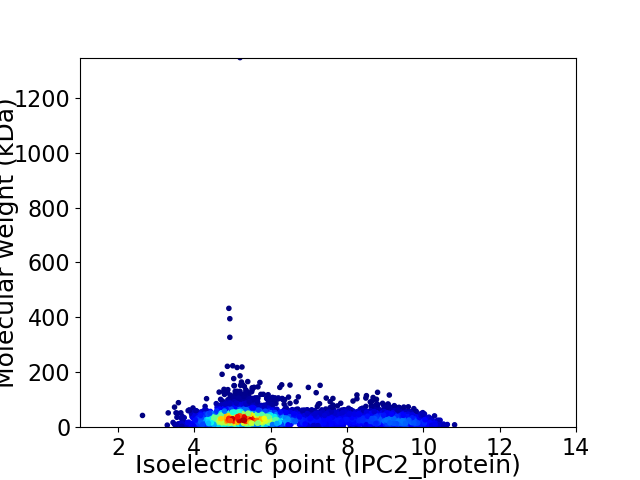
Virtual 2D-PAGE plot for 5294 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X2CET8|A0A1X2CET8_9MYCO TetR family transcriptional regulator OS=Mycobacterium saskatchewanense OX=220927 GN=AWC23_05195 PE=4 SV=1
MM1 pKa = 7.13ITTSGSGPRR10 pKa = 11.84VEE12 pKa = 4.19EE13 pKa = 3.87TSMSFLTAAPEE24 pKa = 4.79AITAAAGNLAHH35 pKa = 7.08IGSSLQEE42 pKa = 3.68ATAAASGQTTTLAAAAADD60 pKa = 4.04EE61 pKa = 4.33VSVAISQLFGTFGEE75 pKa = 4.55DD76 pKa = 3.46FQALSAQAAAFHH88 pKa = 6.27SAFVSVLNGGSAAYY102 pKa = 10.25LSTEE106 pKa = 3.97AASAEE111 pKa = 4.04QVLFHH116 pKa = 7.05AGAAGVPAATLPIIGDD132 pKa = 3.65LGGILGGGTGGTTGGLLGGLGPILSGSPLGPILNGVGQDD171 pKa = 3.43LGGVLSGILGGSPVSLLANPLGPVLQTLSGAFTDD205 pKa = 4.15VPGLQGLGALLQPLLQPVLPALFAPGALPSPAGDD239 pKa = 3.09PWALLFAQTGANLQNLYY256 pKa = 9.1ATWSADD262 pKa = 3.28PFPFLHH268 pKa = 6.56QILVNQQGYY277 pKa = 8.74AQQLGSQMAFALQNFPTTLANAPASLQLGVQSALNFNPVAAAQVFVNQQLGWAGTITTSLQKK339 pKa = 10.79AGADD343 pKa = 3.61LQTTLPVFQADD354 pKa = 3.45MGLASQAIAAGDD366 pKa = 3.55YY367 pKa = 10.51HH368 pKa = 7.23GAVQDD373 pKa = 4.09FTHH376 pKa = 6.76GLLGLFVSGFDD387 pKa = 3.57TSNLSNITVLGPGGDD402 pKa = 4.29LLPILAIPAQQAQAFAGLFPPGSIPGQIAHH432 pKa = 6.81NYY434 pKa = 8.34LNAVSTLTSSTTSTTFGLNLFNPSANVLQADD465 pKa = 4.09AFFGTPLSLAFSVLGAPVSGLNGLATGATVLSAALQSGNGAAAAGALFDD514 pKa = 3.79MPAYY518 pKa = 10.05GLNGFLNGEE527 pKa = 4.51TIVDD531 pKa = 4.24LALPVSTSTVLPSFLGPLGQPLAAILGVAGIDD563 pKa = 3.29PTIPIVIHH571 pKa = 6.35LPFDD575 pKa = 3.88GLLVPPHH582 pKa = 7.06PISSTVDD589 pKa = 3.22VTVGGVLNLPISLTLGGTPFGGLLPLLVNDD619 pKa = 5.65LPRR622 pKa = 11.84QLATAITPP630 pKa = 3.83
MM1 pKa = 7.13ITTSGSGPRR10 pKa = 11.84VEE12 pKa = 4.19EE13 pKa = 3.87TSMSFLTAAPEE24 pKa = 4.79AITAAAGNLAHH35 pKa = 7.08IGSSLQEE42 pKa = 3.68ATAAASGQTTTLAAAAADD60 pKa = 4.04EE61 pKa = 4.33VSVAISQLFGTFGEE75 pKa = 4.55DD76 pKa = 3.46FQALSAQAAAFHH88 pKa = 6.27SAFVSVLNGGSAAYY102 pKa = 10.25LSTEE106 pKa = 3.97AASAEE111 pKa = 4.04QVLFHH116 pKa = 7.05AGAAGVPAATLPIIGDD132 pKa = 3.65LGGILGGGTGGTTGGLLGGLGPILSGSPLGPILNGVGQDD171 pKa = 3.43LGGVLSGILGGSPVSLLANPLGPVLQTLSGAFTDD205 pKa = 4.15VPGLQGLGALLQPLLQPVLPALFAPGALPSPAGDD239 pKa = 3.09PWALLFAQTGANLQNLYY256 pKa = 9.1ATWSADD262 pKa = 3.28PFPFLHH268 pKa = 6.56QILVNQQGYY277 pKa = 8.74AQQLGSQMAFALQNFPTTLANAPASLQLGVQSALNFNPVAAAQVFVNQQLGWAGTITTSLQKK339 pKa = 10.79AGADD343 pKa = 3.61LQTTLPVFQADD354 pKa = 3.45MGLASQAIAAGDD366 pKa = 3.55YY367 pKa = 10.51HH368 pKa = 7.23GAVQDD373 pKa = 4.09FTHH376 pKa = 6.76GLLGLFVSGFDD387 pKa = 3.57TSNLSNITVLGPGGDD402 pKa = 4.29LLPILAIPAQQAQAFAGLFPPGSIPGQIAHH432 pKa = 6.81NYY434 pKa = 8.34LNAVSTLTSSTTSTTFGLNLFNPSANVLQADD465 pKa = 4.09AFFGTPLSLAFSVLGAPVSGLNGLATGATVLSAALQSGNGAAAAGALFDD514 pKa = 3.79MPAYY518 pKa = 10.05GLNGFLNGEE527 pKa = 4.51TIVDD531 pKa = 4.24LALPVSTSTVLPSFLGPLGQPLAAILGVAGIDD563 pKa = 3.29PTIPIVIHH571 pKa = 6.35LPFDD575 pKa = 3.88GLLVPPHH582 pKa = 7.06PISSTVDD589 pKa = 3.22VTVGGVLNLPISLTLGGTPFGGLLPLLVNDD619 pKa = 5.65LPRR622 pKa = 11.84QLATAITPP630 pKa = 3.83
Molecular weight: 62.48 kDa
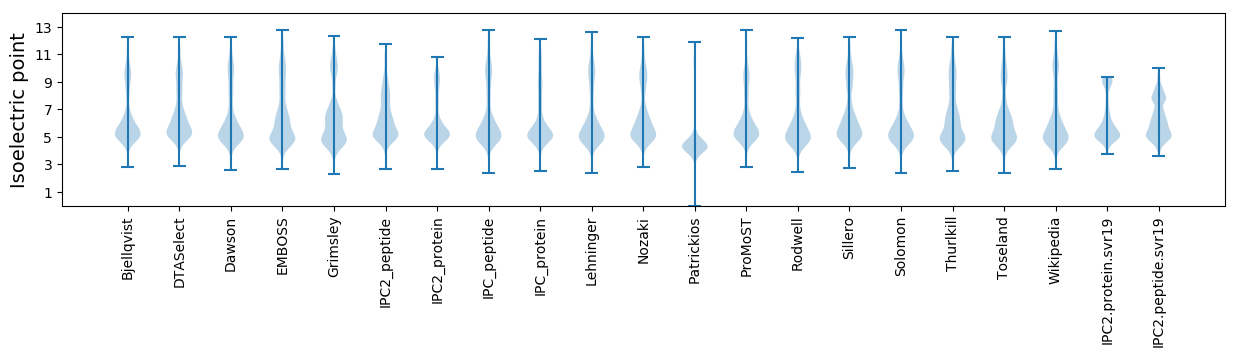
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X2C690|A0A1X2C690_9MYCO Precorrin 6A synthase OS=Mycobacterium saskatchewanense OX=220927 GN=cobF PE=3 SV=1
MM1 pKa = 7.08TALAPTLQAFFTEE14 pKa = 4.29RR15 pKa = 11.84LARR18 pKa = 11.84QKK20 pKa = 10.45NASPHH25 pKa = 4.8TVASYY30 pKa = 11.24RR31 pKa = 11.84DD32 pKa = 3.77TLRR35 pKa = 11.84LLLGFVAEE43 pKa = 4.46RR44 pKa = 11.84TGTPPARR51 pKa = 11.84MQIEE55 pKa = 4.66DD56 pKa = 4.41LDD58 pKa = 4.78APLIGAFLDD67 pKa = 3.29HH68 pKa = 7.52LEE70 pKa = 4.09YY71 pKa = 10.75QRR73 pKa = 11.84ANSVRR78 pKa = 11.84SRR80 pKa = 11.84NTRR83 pKa = 11.84LAAIHH88 pKa = 5.86SLFRR92 pKa = 11.84FAALQHH98 pKa = 6.67PEE100 pKa = 3.76HH101 pKa = 6.86AALIARR107 pKa = 11.84VLAIPPKK114 pKa = 7.9RR115 pKa = 11.84CNRR118 pKa = 11.84AVVSFLNDD126 pKa = 3.25QEE128 pKa = 4.4IDD130 pKa = 3.65ALLAAPDD137 pKa = 3.52TSRR140 pKa = 11.84WIGRR144 pKa = 11.84RR145 pKa = 11.84DD146 pKa = 3.29HH147 pKa = 7.02ALLLVAVQTGLRR159 pKa = 11.84VSEE162 pKa = 4.29LTGLCCGDD170 pKa = 3.35VHH172 pKa = 8.12LGAGAHH178 pKa = 5.1VRR180 pKa = 11.84CVGKK184 pKa = 10.4GRR186 pKa = 11.84KK187 pKa = 7.63QRR189 pKa = 11.84CTPLTAEE196 pKa = 4.33TVAVLRR202 pKa = 11.84GWLAEE207 pKa = 3.86RR208 pKa = 11.84RR209 pKa = 11.84GRR211 pKa = 11.84PEE213 pKa = 5.13DD214 pKa = 3.47PLFPTSRR221 pKa = 11.84GRR223 pKa = 11.84PLSRR227 pKa = 11.84DD228 pKa = 3.18AVALLVTNHH237 pKa = 7.5AITARR242 pKa = 11.84QRR244 pKa = 11.84CPSIDD249 pKa = 3.3AKK251 pKa = 10.48TITPHH256 pKa = 5.29VLRR259 pKa = 11.84HH260 pKa = 4.73STAMTLLRR268 pKa = 11.84AGVDD272 pKa = 3.38TSVIALWLGHH282 pKa = 6.33EE283 pKa = 4.5KK284 pKa = 10.13TDD286 pKa = 3.4TVQIYY291 pKa = 9.11LHH293 pKa = 6.85ADD295 pKa = 3.0LRR297 pKa = 11.84LKK299 pKa = 10.38EE300 pKa = 3.88RR301 pKa = 11.84ALARR305 pKa = 11.84TTPVHH310 pKa = 5.75ATPGRR315 pKa = 11.84YY316 pKa = 8.78RR317 pKa = 11.84PPDD320 pKa = 3.32TLLAFLEE327 pKa = 4.4ALL329 pKa = 3.95
MM1 pKa = 7.08TALAPTLQAFFTEE14 pKa = 4.29RR15 pKa = 11.84LARR18 pKa = 11.84QKK20 pKa = 10.45NASPHH25 pKa = 4.8TVASYY30 pKa = 11.24RR31 pKa = 11.84DD32 pKa = 3.77TLRR35 pKa = 11.84LLLGFVAEE43 pKa = 4.46RR44 pKa = 11.84TGTPPARR51 pKa = 11.84MQIEE55 pKa = 4.66DD56 pKa = 4.41LDD58 pKa = 4.78APLIGAFLDD67 pKa = 3.29HH68 pKa = 7.52LEE70 pKa = 4.09YY71 pKa = 10.75QRR73 pKa = 11.84ANSVRR78 pKa = 11.84SRR80 pKa = 11.84NTRR83 pKa = 11.84LAAIHH88 pKa = 5.86SLFRR92 pKa = 11.84FAALQHH98 pKa = 6.67PEE100 pKa = 3.76HH101 pKa = 6.86AALIARR107 pKa = 11.84VLAIPPKK114 pKa = 7.9RR115 pKa = 11.84CNRR118 pKa = 11.84AVVSFLNDD126 pKa = 3.25QEE128 pKa = 4.4IDD130 pKa = 3.65ALLAAPDD137 pKa = 3.52TSRR140 pKa = 11.84WIGRR144 pKa = 11.84RR145 pKa = 11.84DD146 pKa = 3.29HH147 pKa = 7.02ALLLVAVQTGLRR159 pKa = 11.84VSEE162 pKa = 4.29LTGLCCGDD170 pKa = 3.35VHH172 pKa = 8.12LGAGAHH178 pKa = 5.1VRR180 pKa = 11.84CVGKK184 pKa = 10.4GRR186 pKa = 11.84KK187 pKa = 7.63QRR189 pKa = 11.84CTPLTAEE196 pKa = 4.33TVAVLRR202 pKa = 11.84GWLAEE207 pKa = 3.86RR208 pKa = 11.84RR209 pKa = 11.84GRR211 pKa = 11.84PEE213 pKa = 5.13DD214 pKa = 3.47PLFPTSRR221 pKa = 11.84GRR223 pKa = 11.84PLSRR227 pKa = 11.84DD228 pKa = 3.18AVALLVTNHH237 pKa = 7.5AITARR242 pKa = 11.84QRR244 pKa = 11.84CPSIDD249 pKa = 3.3AKK251 pKa = 10.48TITPHH256 pKa = 5.29VLRR259 pKa = 11.84HH260 pKa = 4.73STAMTLLRR268 pKa = 11.84AGVDD272 pKa = 3.38TSVIALWLGHH282 pKa = 6.33EE283 pKa = 4.5KK284 pKa = 10.13TDD286 pKa = 3.4TVQIYY291 pKa = 9.11LHH293 pKa = 6.85ADD295 pKa = 3.0LRR297 pKa = 11.84LKK299 pKa = 10.38EE300 pKa = 3.88RR301 pKa = 11.84ALARR305 pKa = 11.84TTPVHH310 pKa = 5.75ATPGRR315 pKa = 11.84YY316 pKa = 8.78RR317 pKa = 11.84PPDD320 pKa = 3.32TLLAFLEE327 pKa = 4.4ALL329 pKa = 3.95
Molecular weight: 36.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1722884 |
27 |
12626 |
325.4 |
34.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.26 ± 0.052 | 0.832 ± 0.011 |
6.151 ± 0.031 | 5.168 ± 0.032 |
3.058 ± 0.02 | 9.107 ± 0.039 |
2.258 ± 0.016 | 4.118 ± 0.023 |
2.097 ± 0.02 | 9.861 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.02 ± 0.014 | 2.248 ± 0.018 |
5.988 ± 0.029 | 2.955 ± 0.019 |
7.507 ± 0.037 | 5.37 ± 0.025 |
5.76 ± 0.023 | 8.626 ± 0.036 |
1.474 ± 0.014 | 2.143 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
