
Staphylococcus phage IME1323_01
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Azeredovirinae; unclassified Azeredovirinae
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
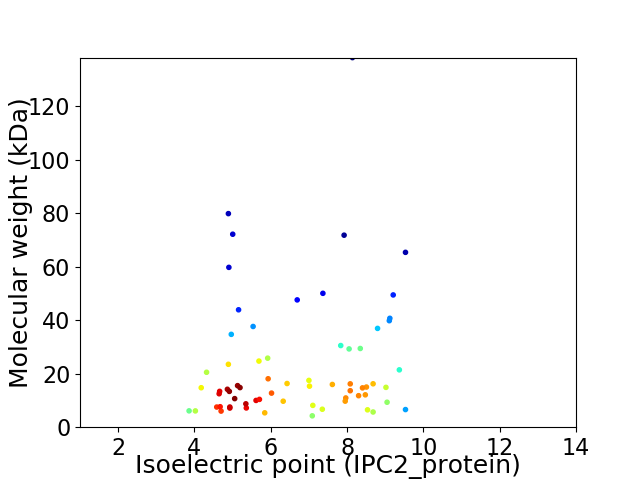
Virtual 2D-PAGE plot for 67 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6JNQ0|A0A1W6JNQ0_9CAUD ATPase OS=Staphylococcus phage IME1323_01 OX=1965487 PE=4 SV=1
MM1 pKa = 7.22VKK3 pKa = 9.83IKK5 pKa = 10.23EE6 pKa = 4.11IQTDD10 pKa = 3.92GTYY13 pKa = 10.24TIYY16 pKa = 10.97DD17 pKa = 3.8SNDD20 pKa = 2.65NSINITKK27 pKa = 10.6EE28 pKa = 3.45EE29 pKa = 3.98FEE31 pKa = 4.7EE32 pKa = 4.12IKK34 pKa = 10.59QYY36 pKa = 11.19SIDD39 pKa = 3.56SEE41 pKa = 5.01YY42 pKa = 11.5NFLSSEE48 pKa = 4.13DD49 pKa = 3.75TQAA52 pKa = 4.25
MM1 pKa = 7.22VKK3 pKa = 9.83IKK5 pKa = 10.23EE6 pKa = 4.11IQTDD10 pKa = 3.92GTYY13 pKa = 10.24TIYY16 pKa = 10.97DD17 pKa = 3.8SNDD20 pKa = 2.65NSINITKK27 pKa = 10.6EE28 pKa = 3.45EE29 pKa = 3.98FEE31 pKa = 4.7EE32 pKa = 4.12IKK34 pKa = 10.59QYY36 pKa = 11.19SIDD39 pKa = 3.56SEE41 pKa = 5.01YY42 pKa = 11.5NFLSSEE48 pKa = 4.13DD49 pKa = 3.75TQAA52 pKa = 4.25
Molecular weight: 6.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6JNR2|A0A1W6JNR2_9CAUD Uncharacterized protein OS=Staphylococcus phage IME1323_01 OX=1965487 PE=4 SV=1
MM1 pKa = 7.44AQRR4 pKa = 11.84NNYY7 pKa = 8.54PNSSWKK13 pKa = 9.98IYY15 pKa = 10.41RR16 pKa = 11.84NTPSFVPKK24 pKa = 10.4PGDD27 pKa = 3.52IVCWTYY33 pKa = 11.15GWAGHH38 pKa = 5.53TGIVVGPSDD47 pKa = 3.58KK48 pKa = 10.35KK49 pKa = 8.97TFRR52 pKa = 11.84TVDD55 pKa = 3.57QNWFGANQYY64 pKa = 9.97SGSRR68 pKa = 11.84AAYY71 pKa = 9.65VKK73 pKa = 9.34HH74 pKa = 6.33TYY76 pKa = 10.9NGMGGNIYY84 pKa = 10.25FIRR87 pKa = 11.84PPYY90 pKa = 9.4KK91 pKa = 10.31ASNNSAPAKK100 pKa = 10.33KK101 pKa = 9.76QDD103 pKa = 3.51TNKK106 pKa = 9.64VVTKK110 pKa = 9.91KK111 pKa = 10.5KK112 pKa = 10.25KK113 pKa = 8.07RR114 pKa = 11.84TEE116 pKa = 3.51ISFTIDD122 pKa = 3.29DD123 pKa = 4.43NEE125 pKa = 4.38TIYY128 pKa = 10.87PEE130 pKa = 4.83FIPHH134 pKa = 6.56QIVLGNDD141 pKa = 3.09RR142 pKa = 11.84GHH144 pKa = 5.99SPKK147 pKa = 10.78GVTVRR152 pKa = 11.84NAGTMCSVQDD162 pKa = 3.71MYY164 pKa = 11.44FDD166 pKa = 3.47RR167 pKa = 11.84QKK169 pKa = 10.92YY170 pKa = 9.96IKK172 pKa = 10.38DD173 pKa = 3.26KK174 pKa = 10.54EE175 pKa = 4.29YY176 pKa = 9.64PHH178 pKa = 6.96YY179 pKa = 10.66FIDD182 pKa = 5.64RR183 pKa = 11.84NHH185 pKa = 6.03IWQPRR190 pKa = 11.84LEE192 pKa = 4.41TYY194 pKa = 8.16EE195 pKa = 4.37VPSDD199 pKa = 3.7PQNLVIEE206 pKa = 4.56VCGDD210 pKa = 3.47LSDD213 pKa = 4.43SKK215 pKa = 11.82NDD217 pKa = 4.09FILNEE222 pKa = 3.93LHH224 pKa = 7.26AMIFVTQRR232 pKa = 11.84MKK234 pKa = 10.72FHH236 pKa = 6.92NISLKK241 pKa = 10.44KK242 pKa = 10.58SSINVKK248 pKa = 10.42DD249 pKa = 3.66PVWRR253 pKa = 11.84SIYY256 pKa = 10.5EE257 pKa = 4.04HH258 pKa = 7.09GDD260 pKa = 2.65WNTALKK266 pKa = 10.64GKK268 pKa = 10.02APKK271 pKa = 10.04KK272 pKa = 10.69VIDD275 pKa = 3.8KK276 pKa = 9.7TVEE279 pKa = 3.54ALMYY283 pKa = 10.45LYY285 pKa = 10.68GHH287 pKa = 7.13RR288 pKa = 11.84KK289 pKa = 7.16TLLTEE294 pKa = 4.07IPRR297 pKa = 11.84DD298 pKa = 3.57KK299 pKa = 10.36ITTRR303 pKa = 11.84TIRR306 pKa = 11.84VSVPSSSSKK315 pKa = 10.72NSNSTSSSSKK325 pKa = 10.68KK326 pKa = 10.4NSTSHH331 pKa = 6.21NSSHH335 pKa = 6.05STTKK339 pKa = 10.42KK340 pKa = 10.54KK341 pKa = 9.45KK342 pKa = 6.19TTNKK346 pKa = 9.66KK347 pKa = 7.93QNQVIVEE354 pKa = 3.97HH355 pKa = 6.4SKK357 pKa = 9.14YY358 pKa = 9.58TFNQAVNIQSRR369 pKa = 11.84LSPQINYY376 pKa = 9.26GVGWYY381 pKa = 9.51NATRR385 pKa = 11.84SQTLNAMNSLQIWNSGTQKK404 pKa = 10.8YY405 pKa = 9.08QMLNLGKK412 pKa = 9.63YY413 pKa = 9.45QGISVTALNKK423 pKa = 9.62ILRR426 pKa = 11.84GKK428 pKa = 10.68GSLSGQGKK436 pKa = 9.31AVAYY440 pKa = 9.94ACKK443 pKa = 10.06KK444 pKa = 9.61YY445 pKa = 10.83NLNEE449 pKa = 3.84IYY451 pKa = 10.47LISHH455 pKa = 7.21AFLEE459 pKa = 4.57SGYY462 pKa = 9.98GRR464 pKa = 11.84SYY466 pKa = 10.73FSSGRR471 pKa = 11.84AGVYY475 pKa = 10.16NYY477 pKa = 10.44FGIGAFDD484 pKa = 4.28SNPNNAINYY493 pKa = 7.55ARR495 pKa = 11.84SHH497 pKa = 5.71GWTTPSKK504 pKa = 10.76GIIGGARR511 pKa = 11.84FVRR514 pKa = 11.84RR515 pKa = 11.84GYY517 pKa = 10.18ISQGQNTLYY526 pKa = 10.78RR527 pKa = 11.84MRR529 pKa = 11.84WNPKK533 pKa = 9.65HH534 pKa = 6.64PGTHH538 pKa = 5.68QYY540 pKa = 9.23ATDD543 pKa = 3.61VRR545 pKa = 11.84WAQVQATTIKK555 pKa = 10.49SLYY558 pKa = 7.82EE559 pKa = 3.58QIGIKK564 pKa = 10.4GEE566 pKa = 3.79YY567 pKa = 9.66FIRR570 pKa = 11.84DD571 pKa = 3.27RR572 pKa = 11.84YY573 pKa = 9.4KK574 pKa = 10.69
MM1 pKa = 7.44AQRR4 pKa = 11.84NNYY7 pKa = 8.54PNSSWKK13 pKa = 9.98IYY15 pKa = 10.41RR16 pKa = 11.84NTPSFVPKK24 pKa = 10.4PGDD27 pKa = 3.52IVCWTYY33 pKa = 11.15GWAGHH38 pKa = 5.53TGIVVGPSDD47 pKa = 3.58KK48 pKa = 10.35KK49 pKa = 8.97TFRR52 pKa = 11.84TVDD55 pKa = 3.57QNWFGANQYY64 pKa = 9.97SGSRR68 pKa = 11.84AAYY71 pKa = 9.65VKK73 pKa = 9.34HH74 pKa = 6.33TYY76 pKa = 10.9NGMGGNIYY84 pKa = 10.25FIRR87 pKa = 11.84PPYY90 pKa = 9.4KK91 pKa = 10.31ASNNSAPAKK100 pKa = 10.33KK101 pKa = 9.76QDD103 pKa = 3.51TNKK106 pKa = 9.64VVTKK110 pKa = 9.91KK111 pKa = 10.5KK112 pKa = 10.25KK113 pKa = 8.07RR114 pKa = 11.84TEE116 pKa = 3.51ISFTIDD122 pKa = 3.29DD123 pKa = 4.43NEE125 pKa = 4.38TIYY128 pKa = 10.87PEE130 pKa = 4.83FIPHH134 pKa = 6.56QIVLGNDD141 pKa = 3.09RR142 pKa = 11.84GHH144 pKa = 5.99SPKK147 pKa = 10.78GVTVRR152 pKa = 11.84NAGTMCSVQDD162 pKa = 3.71MYY164 pKa = 11.44FDD166 pKa = 3.47RR167 pKa = 11.84QKK169 pKa = 10.92YY170 pKa = 9.96IKK172 pKa = 10.38DD173 pKa = 3.26KK174 pKa = 10.54EE175 pKa = 4.29YY176 pKa = 9.64PHH178 pKa = 6.96YY179 pKa = 10.66FIDD182 pKa = 5.64RR183 pKa = 11.84NHH185 pKa = 6.03IWQPRR190 pKa = 11.84LEE192 pKa = 4.41TYY194 pKa = 8.16EE195 pKa = 4.37VPSDD199 pKa = 3.7PQNLVIEE206 pKa = 4.56VCGDD210 pKa = 3.47LSDD213 pKa = 4.43SKK215 pKa = 11.82NDD217 pKa = 4.09FILNEE222 pKa = 3.93LHH224 pKa = 7.26AMIFVTQRR232 pKa = 11.84MKK234 pKa = 10.72FHH236 pKa = 6.92NISLKK241 pKa = 10.44KK242 pKa = 10.58SSINVKK248 pKa = 10.42DD249 pKa = 3.66PVWRR253 pKa = 11.84SIYY256 pKa = 10.5EE257 pKa = 4.04HH258 pKa = 7.09GDD260 pKa = 2.65WNTALKK266 pKa = 10.64GKK268 pKa = 10.02APKK271 pKa = 10.04KK272 pKa = 10.69VIDD275 pKa = 3.8KK276 pKa = 9.7TVEE279 pKa = 3.54ALMYY283 pKa = 10.45LYY285 pKa = 10.68GHH287 pKa = 7.13RR288 pKa = 11.84KK289 pKa = 7.16TLLTEE294 pKa = 4.07IPRR297 pKa = 11.84DD298 pKa = 3.57KK299 pKa = 10.36ITTRR303 pKa = 11.84TIRR306 pKa = 11.84VSVPSSSSKK315 pKa = 10.72NSNSTSSSSKK325 pKa = 10.68KK326 pKa = 10.4NSTSHH331 pKa = 6.21NSSHH335 pKa = 6.05STTKK339 pKa = 10.42KK340 pKa = 10.54KK341 pKa = 9.45KK342 pKa = 6.19TTNKK346 pKa = 9.66KK347 pKa = 7.93QNQVIVEE354 pKa = 3.97HH355 pKa = 6.4SKK357 pKa = 9.14YY358 pKa = 9.58TFNQAVNIQSRR369 pKa = 11.84LSPQINYY376 pKa = 9.26GVGWYY381 pKa = 9.51NATRR385 pKa = 11.84SQTLNAMNSLQIWNSGTQKK404 pKa = 10.8YY405 pKa = 9.08QMLNLGKK412 pKa = 9.63YY413 pKa = 9.45QGISVTALNKK423 pKa = 9.62ILRR426 pKa = 11.84GKK428 pKa = 10.68GSLSGQGKK436 pKa = 9.31AVAYY440 pKa = 9.94ACKK443 pKa = 10.06KK444 pKa = 9.61YY445 pKa = 10.83NLNEE449 pKa = 3.84IYY451 pKa = 10.47LISHH455 pKa = 7.21AFLEE459 pKa = 4.57SGYY462 pKa = 9.98GRR464 pKa = 11.84SYY466 pKa = 10.73FSSGRR471 pKa = 11.84AGVYY475 pKa = 10.16NYY477 pKa = 10.44FGIGAFDD484 pKa = 4.28SNPNNAINYY493 pKa = 7.55ARR495 pKa = 11.84SHH497 pKa = 5.71GWTTPSKK504 pKa = 10.76GIIGGARR511 pKa = 11.84FVRR514 pKa = 11.84RR515 pKa = 11.84GYY517 pKa = 10.18ISQGQNTLYY526 pKa = 10.78RR527 pKa = 11.84MRR529 pKa = 11.84WNPKK533 pKa = 9.65HH534 pKa = 6.64PGTHH538 pKa = 5.68QYY540 pKa = 9.23ATDD543 pKa = 3.61VRR545 pKa = 11.84WAQVQATTIKK555 pKa = 10.49SLYY558 pKa = 7.82EE559 pKa = 3.58QIGIKK564 pKa = 10.4GEE566 pKa = 3.79YY567 pKa = 9.66FIRR570 pKa = 11.84DD571 pKa = 3.27RR572 pKa = 11.84YY573 pKa = 9.4KK574 pKa = 10.69
Molecular weight: 65.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13645 |
38 |
1302 |
203.7 |
23.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.738 ± 0.808 | 0.652 ± 0.109 |
6.259 ± 0.35 | 7.131 ± 0.476 |
4.295 ± 0.201 | 5.694 ± 0.356 |
1.979 ± 0.228 | 7.534 ± 0.32 |
9.154 ± 0.474 | 7.453 ± 0.277 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.426 ± 0.182 | 7.028 ± 0.394 |
2.682 ± 0.205 | 3.694 ± 0.194 |
4.075 ± 0.326 | 6.207 ± 0.309 |
6.083 ± 0.332 | 6.31 ± 0.375 |
1.217 ± 0.151 | 4.39 ± 0.419 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
