
Methanohalobium evestigatum (strain ATCC BAA-1072 / DSM 3721 / NBRC 107634 / OCM 161 / Z-7303)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanosarcinales; Methanosarcinaceae; Methanohalobium; Methanohalobium evestigatum
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
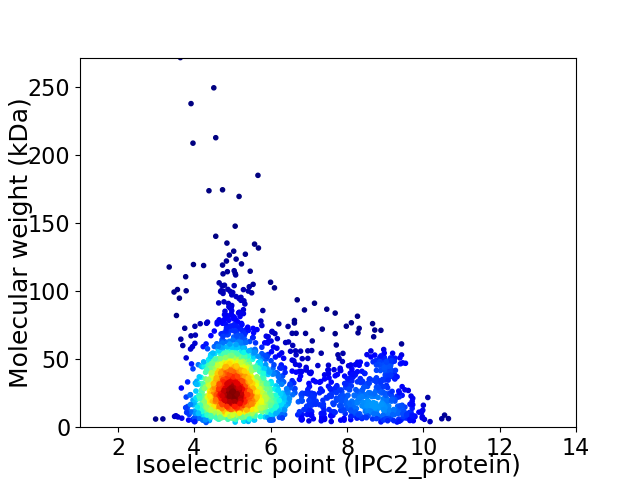
Virtual 2D-PAGE plot for 2250 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7E9Z3|D7E9Z3_METEZ HTH_45 domain-containing protein OS=Methanohalobium evestigatum (strain ATCC BAA-1072 / DSM 3721 / NBRC 107634 / OCM 161 / Z-7303) OX=644295 GN=Metev_1568 PE=4 SV=1
MM1 pKa = 7.29KK2 pKa = 10.55RR3 pKa = 11.84LMVLFAGLMAILMSISAASAITVDD27 pKa = 3.77GNKK30 pKa = 9.58TSGEE34 pKa = 3.9WDD36 pKa = 3.61EE37 pKa = 4.55NWAFGQTNNATAASEE52 pKa = 3.95YY53 pKa = 10.72DD54 pKa = 3.4INNLGDD60 pKa = 3.79RR61 pKa = 11.84LEE63 pKa = 4.3IMQGTLQADD72 pKa = 3.57TGDD75 pKa = 3.99YY76 pKa = 10.97NAVDD80 pKa = 4.27PKK82 pKa = 10.88NDD84 pKa = 3.5SGSSYY89 pKa = 11.01AEE91 pKa = 3.75SMALNGNSSGSDD103 pKa = 3.15LYY105 pKa = 11.08KK106 pKa = 10.87VYY108 pKa = 10.57GHH110 pKa = 6.29YY111 pKa = 10.34NQSTDD116 pKa = 2.92TLYY119 pKa = 11.53GMTEE123 pKa = 4.21VYY125 pKa = 10.4GIPGDD130 pKa = 3.55HH131 pKa = 7.55DD132 pKa = 4.56GNGDD136 pKa = 3.58VTTEE140 pKa = 3.98SSAGDD145 pKa = 3.22SGGDD149 pKa = 3.27VGPAGFGLSTQEE161 pKa = 3.55SWSIAFYY168 pKa = 9.28QVKK171 pKa = 10.57NGDD174 pKa = 3.68YY175 pKa = 10.29QDD177 pKa = 3.8YY178 pKa = 11.2SLIQISDD185 pKa = 3.86NDD187 pKa = 3.8WNIIDD192 pKa = 4.14TDD194 pKa = 3.62VGLDD198 pKa = 3.64YY199 pKa = 11.54DD200 pKa = 3.91NVSAKK205 pKa = 10.34FSYY208 pKa = 10.86QNFRR212 pKa = 11.84QIGDD216 pKa = 3.46DD217 pKa = 3.7TLPKK221 pKa = 9.87SVYY224 pKa = 9.39EE225 pKa = 3.87IKK227 pKa = 10.78VEE229 pKa = 4.06NFSKK233 pKa = 10.66FYY235 pKa = 10.91DD236 pKa = 3.66VNPGSKK242 pKa = 10.22LAIQVGAGSNGDD254 pKa = 3.66PQVGEE259 pKa = 4.02DD260 pKa = 3.33SGVVFFEE267 pKa = 4.46IPNPQIDD274 pKa = 3.56IEE276 pKa = 4.6KK277 pKa = 8.79FTKK280 pKa = 10.67DD281 pKa = 3.95MGGDD285 pKa = 3.19WQQADD290 pKa = 3.54NSADD294 pKa = 3.53PDD296 pKa = 3.89VPFFNSAGQTVEE308 pKa = 3.5WKK310 pKa = 11.25YY311 pKa = 11.11EE312 pKa = 3.77VTNNGTVPFDD322 pKa = 3.69LANVTVTDD330 pKa = 4.36DD331 pKa = 3.62KK332 pKa = 11.71GVTPVLDD339 pKa = 3.67EE340 pKa = 4.56TSDD343 pKa = 4.29DD344 pKa = 4.04GNDD347 pKa = 4.56DD348 pKa = 3.46ILSPGEE354 pKa = 3.91TWTYY358 pKa = 7.71TASGTVEE365 pKa = 4.09EE366 pKa = 4.8CPEE369 pKa = 4.17GSVYY373 pKa = 10.28TYY375 pKa = 11.32EE376 pKa = 5.03NIGTVNAISPFGEE389 pKa = 4.35VNDD392 pKa = 4.79TDD394 pKa = 3.99PSHH397 pKa = 6.71YY398 pKa = 9.97KK399 pKa = 10.03CQEE402 pKa = 3.71PTPGIDD408 pKa = 2.97IEE410 pKa = 4.85KK411 pKa = 8.48YY412 pKa = 8.99TNGEE416 pKa = 4.04NADD419 pKa = 3.82TGPGPKK425 pKa = 9.66VSNEE429 pKa = 3.95STVTWTYY436 pKa = 10.17EE437 pKa = 3.65VHH439 pKa = 6.71NNGSDD444 pKa = 3.48DD445 pKa = 3.91LQNVQVTDD453 pKa = 4.22DD454 pKa = 3.88KK455 pKa = 11.65LGTLPEE461 pKa = 4.0TAIVDD466 pKa = 3.89KK467 pKa = 11.43GDD469 pKa = 3.75GDD471 pKa = 5.06DD472 pKa = 3.94ILNIGEE478 pKa = 4.17TWVYY482 pKa = 11.23EE483 pKa = 4.09MTGTADD489 pKa = 4.22CNFTEE494 pKa = 4.79DD495 pKa = 3.79NPYY498 pKa = 11.23EE499 pKa = 4.17NTATTNGTGVITGEE513 pKa = 4.34SVEE516 pKa = 4.44DD517 pKa = 3.85TDD519 pKa = 5.09PSHH522 pKa = 6.91YY523 pKa = 10.14YY524 pKa = 10.22CKK526 pKa = 10.17PPNGVPALTPTSLMGLIGALGLIGIVGLKK555 pKa = 10.28RR556 pKa = 11.84RR557 pKa = 11.84DD558 pKa = 3.25
MM1 pKa = 7.29KK2 pKa = 10.55RR3 pKa = 11.84LMVLFAGLMAILMSISAASAITVDD27 pKa = 3.77GNKK30 pKa = 9.58TSGEE34 pKa = 3.9WDD36 pKa = 3.61EE37 pKa = 4.55NWAFGQTNNATAASEE52 pKa = 3.95YY53 pKa = 10.72DD54 pKa = 3.4INNLGDD60 pKa = 3.79RR61 pKa = 11.84LEE63 pKa = 4.3IMQGTLQADD72 pKa = 3.57TGDD75 pKa = 3.99YY76 pKa = 10.97NAVDD80 pKa = 4.27PKK82 pKa = 10.88NDD84 pKa = 3.5SGSSYY89 pKa = 11.01AEE91 pKa = 3.75SMALNGNSSGSDD103 pKa = 3.15LYY105 pKa = 11.08KK106 pKa = 10.87VYY108 pKa = 10.57GHH110 pKa = 6.29YY111 pKa = 10.34NQSTDD116 pKa = 2.92TLYY119 pKa = 11.53GMTEE123 pKa = 4.21VYY125 pKa = 10.4GIPGDD130 pKa = 3.55HH131 pKa = 7.55DD132 pKa = 4.56GNGDD136 pKa = 3.58VTTEE140 pKa = 3.98SSAGDD145 pKa = 3.22SGGDD149 pKa = 3.27VGPAGFGLSTQEE161 pKa = 3.55SWSIAFYY168 pKa = 9.28QVKK171 pKa = 10.57NGDD174 pKa = 3.68YY175 pKa = 10.29QDD177 pKa = 3.8YY178 pKa = 11.2SLIQISDD185 pKa = 3.86NDD187 pKa = 3.8WNIIDD192 pKa = 4.14TDD194 pKa = 3.62VGLDD198 pKa = 3.64YY199 pKa = 11.54DD200 pKa = 3.91NVSAKK205 pKa = 10.34FSYY208 pKa = 10.86QNFRR212 pKa = 11.84QIGDD216 pKa = 3.46DD217 pKa = 3.7TLPKK221 pKa = 9.87SVYY224 pKa = 9.39EE225 pKa = 3.87IKK227 pKa = 10.78VEE229 pKa = 4.06NFSKK233 pKa = 10.66FYY235 pKa = 10.91DD236 pKa = 3.66VNPGSKK242 pKa = 10.22LAIQVGAGSNGDD254 pKa = 3.66PQVGEE259 pKa = 4.02DD260 pKa = 3.33SGVVFFEE267 pKa = 4.46IPNPQIDD274 pKa = 3.56IEE276 pKa = 4.6KK277 pKa = 8.79FTKK280 pKa = 10.67DD281 pKa = 3.95MGGDD285 pKa = 3.19WQQADD290 pKa = 3.54NSADD294 pKa = 3.53PDD296 pKa = 3.89VPFFNSAGQTVEE308 pKa = 3.5WKK310 pKa = 11.25YY311 pKa = 11.11EE312 pKa = 3.77VTNNGTVPFDD322 pKa = 3.69LANVTVTDD330 pKa = 4.36DD331 pKa = 3.62KK332 pKa = 11.71GVTPVLDD339 pKa = 3.67EE340 pKa = 4.56TSDD343 pKa = 4.29DD344 pKa = 4.04GNDD347 pKa = 4.56DD348 pKa = 3.46ILSPGEE354 pKa = 3.91TWTYY358 pKa = 7.71TASGTVEE365 pKa = 4.09EE366 pKa = 4.8CPEE369 pKa = 4.17GSVYY373 pKa = 10.28TYY375 pKa = 11.32EE376 pKa = 5.03NIGTVNAISPFGEE389 pKa = 4.35VNDD392 pKa = 4.79TDD394 pKa = 3.99PSHH397 pKa = 6.71YY398 pKa = 9.97KK399 pKa = 10.03CQEE402 pKa = 3.71PTPGIDD408 pKa = 2.97IEE410 pKa = 4.85KK411 pKa = 8.48YY412 pKa = 8.99TNGEE416 pKa = 4.04NADD419 pKa = 3.82TGPGPKK425 pKa = 9.66VSNEE429 pKa = 3.95STVTWTYY436 pKa = 10.17EE437 pKa = 3.65VHH439 pKa = 6.71NNGSDD444 pKa = 3.48DD445 pKa = 3.91LQNVQVTDD453 pKa = 4.22DD454 pKa = 3.88KK455 pKa = 11.65LGTLPEE461 pKa = 4.0TAIVDD466 pKa = 3.89KK467 pKa = 11.43GDD469 pKa = 3.75GDD471 pKa = 5.06DD472 pKa = 3.94ILNIGEE478 pKa = 4.17TWVYY482 pKa = 11.23EE483 pKa = 4.09MTGTADD489 pKa = 4.22CNFTEE494 pKa = 4.79DD495 pKa = 3.79NPYY498 pKa = 11.23EE499 pKa = 4.17NTATTNGTGVITGEE513 pKa = 4.34SVEE516 pKa = 4.44DD517 pKa = 3.85TDD519 pKa = 5.09PSHH522 pKa = 6.91YY523 pKa = 10.14YY524 pKa = 10.22CKK526 pKa = 10.17PPNGVPALTPTSLMGLIGALGLIGIVGLKK555 pKa = 10.28RR556 pKa = 11.84RR557 pKa = 11.84DD558 pKa = 3.25
Molecular weight: 60.01 kDa
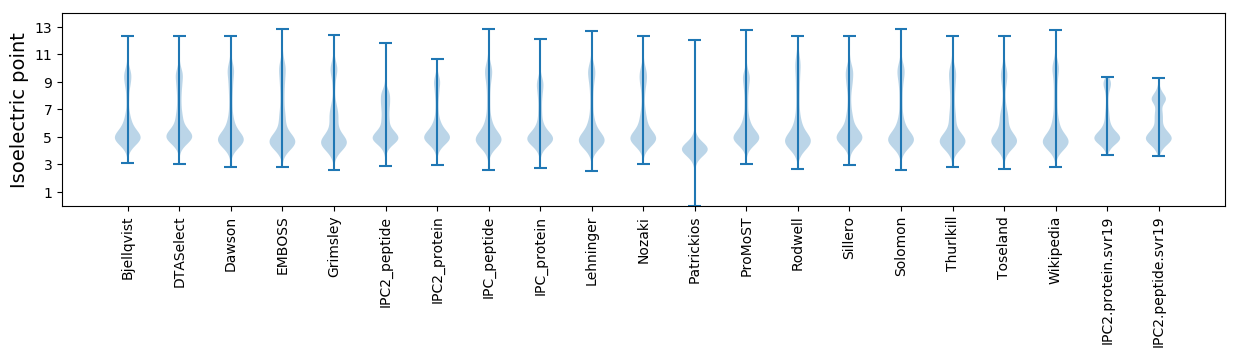
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7E8J1|D7E8J1_METEZ Pilin_N domain-containing protein OS=Methanohalobium evestigatum (strain ATCC BAA-1072 / DSM 3721 / NBRC 107634 / OCM 161 / Z-7303) OX=644295 GN=Metev_0627 PE=4 SV=1
MM1 pKa = 7.55AKK3 pKa = 10.25GYY5 pKa = 7.33RR6 pKa = 11.84TKK8 pKa = 11.17GRR10 pKa = 11.84VTRR13 pKa = 11.84SAGRR17 pKa = 11.84FGTRR21 pKa = 11.84YY22 pKa = 9.33GRR24 pKa = 11.84KK25 pKa = 9.16DD26 pKa = 3.08RR27 pKa = 11.84KK28 pKa = 10.24LIANIEE34 pKa = 4.09EE35 pKa = 4.22QMHH38 pKa = 5.98KK39 pKa = 10.33KK40 pKa = 9.52HH41 pKa = 6.38RR42 pKa = 11.84CSNCARR48 pKa = 11.84VSVRR52 pKa = 11.84RR53 pKa = 11.84TGTGIWEE60 pKa = 4.34CTKK63 pKa = 10.56CGHH66 pKa = 6.1TFAGGTYY73 pKa = 9.32TPQTSVGKK81 pKa = 7.63TVPRR85 pKa = 11.84AIRR88 pKa = 11.84KK89 pKa = 9.27AIEE92 pKa = 4.2EE93 pKa = 4.11EE94 pKa = 4.52VEE96 pKa = 3.91
MM1 pKa = 7.55AKK3 pKa = 10.25GYY5 pKa = 7.33RR6 pKa = 11.84TKK8 pKa = 11.17GRR10 pKa = 11.84VTRR13 pKa = 11.84SAGRR17 pKa = 11.84FGTRR21 pKa = 11.84YY22 pKa = 9.33GRR24 pKa = 11.84KK25 pKa = 9.16DD26 pKa = 3.08RR27 pKa = 11.84KK28 pKa = 10.24LIANIEE34 pKa = 4.09EE35 pKa = 4.22QMHH38 pKa = 5.98KK39 pKa = 10.33KK40 pKa = 9.52HH41 pKa = 6.38RR42 pKa = 11.84CSNCARR48 pKa = 11.84VSVRR52 pKa = 11.84RR53 pKa = 11.84TGTGIWEE60 pKa = 4.34CTKK63 pKa = 10.56CGHH66 pKa = 6.1TFAGGTYY73 pKa = 9.32TPQTSVGKK81 pKa = 7.63TVPRR85 pKa = 11.84AIRR88 pKa = 11.84KK89 pKa = 9.27AIEE92 pKa = 4.2EE93 pKa = 4.11EE94 pKa = 4.52VEE96 pKa = 3.91
Molecular weight: 10.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
666791 |
31 |
2559 |
296.4 |
33.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.654 ± 0.051 | 1.14 ± 0.022 |
6.477 ± 0.053 | 7.358 ± 0.057 |
3.863 ± 0.038 | 6.723 ± 0.05 |
1.83 ± 0.021 | 8.446 ± 0.056 |
7.253 ± 0.066 | 8.466 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.524 ± 0.029 | 5.926 ± 0.07 |
3.597 ± 0.026 | 2.8 ± 0.026 |
3.814 ± 0.041 | 6.937 ± 0.048 |
5.491 ± 0.054 | 6.913 ± 0.038 |
0.849 ± 0.018 | 3.936 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
