
Canis familiaris papillomavirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Lambdapapillomavirus; Lambdapapillomavirus 3
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
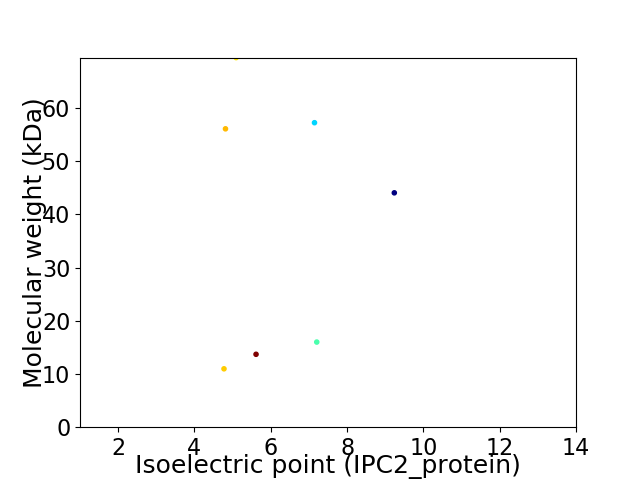
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C8YJL4|C8YJL4_9PAPI Replication protein E1 OS=Canis familiaris papillomavirus 6 OX=1513269 GN=E1 PE=3 SV=1
MM1 pKa = 7.61IGQEE5 pKa = 4.02PTLPDD10 pKa = 3.22IVLTAQLPPEE20 pKa = 4.43TVDD23 pKa = 2.94LHH25 pKa = 8.24CYY27 pKa = 7.4EE28 pKa = 4.6QIPAEE33 pKa = 4.12EE34 pKa = 4.33EE35 pKa = 4.18EE36 pKa = 4.45EE37 pKa = 4.2EE38 pKa = 4.45EE39 pKa = 4.08PASRR43 pKa = 11.84DD44 pKa = 3.55LYY46 pKa = 10.78RR47 pKa = 11.84VAALCGLCGSRR58 pKa = 11.84VRR60 pKa = 11.84FVCLAEE66 pKa = 4.85SDD68 pKa = 4.62DD69 pKa = 3.35IRR71 pKa = 11.84HH72 pKa = 5.56FQRR75 pKa = 11.84LLFNLSFVCLRR86 pKa = 11.84CVKK89 pKa = 9.57TEE91 pKa = 3.85KK92 pKa = 10.49LNHH95 pKa = 6.27GGG97 pKa = 3.44
MM1 pKa = 7.61IGQEE5 pKa = 4.02PTLPDD10 pKa = 3.22IVLTAQLPPEE20 pKa = 4.43TVDD23 pKa = 2.94LHH25 pKa = 8.24CYY27 pKa = 7.4EE28 pKa = 4.6QIPAEE33 pKa = 4.12EE34 pKa = 4.33EE35 pKa = 4.18EE36 pKa = 4.45EE37 pKa = 4.2EE38 pKa = 4.45EE39 pKa = 4.08PASRR43 pKa = 11.84DD44 pKa = 3.55LYY46 pKa = 10.78RR47 pKa = 11.84VAALCGLCGSRR58 pKa = 11.84VRR60 pKa = 11.84FVCLAEE66 pKa = 4.85SDD68 pKa = 4.62DD69 pKa = 3.35IRR71 pKa = 11.84HH72 pKa = 5.56FQRR75 pKa = 11.84LLFNLSFVCLRR86 pKa = 11.84CVKK89 pKa = 9.57TEE91 pKa = 3.85KK92 pKa = 10.49LNHH95 pKa = 6.27GGG97 pKa = 3.44
Molecular weight: 10.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C8YJL6|C8YJL6_9PAPI E4 (Fragment) OS=Canis familiaris papillomavirus 6 OX=1513269 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 5.32NLSKK6 pKa = 10.98ALDD9 pKa = 3.73SLQEE13 pKa = 4.1EE14 pKa = 4.64LLSLYY19 pKa = 9.66EE20 pKa = 4.39KK21 pKa = 10.91NSGDD25 pKa = 3.11IADD28 pKa = 5.35QIRR31 pKa = 11.84HH32 pKa = 4.87WNLVRR37 pKa = 11.84KK38 pKa = 8.51EE39 pKa = 3.68QVLFHH44 pKa = 6.48FARR47 pKa = 11.84KK48 pKa = 9.78NGVSRR53 pKa = 11.84IGMSPVPPQSVSQQKK68 pKa = 10.08AKK70 pKa = 10.69SAIEE74 pKa = 3.9QEE76 pKa = 4.17ILLQSLADD84 pKa = 3.56SQYY87 pKa = 11.0GSEE90 pKa = 4.28RR91 pKa = 11.84WTLSDD96 pKa = 3.23TSKK99 pKa = 10.92EE100 pKa = 3.9RR101 pKa = 11.84LLADD105 pKa = 3.91PPYY108 pKa = 10.54CFKK111 pKa = 11.33KK112 pKa = 10.39NGKK115 pKa = 8.86QVDD118 pKa = 3.59VRR120 pKa = 11.84FDD122 pKa = 3.63EE123 pKa = 4.71QPDD126 pKa = 3.49NLVRR130 pKa = 11.84YY131 pKa = 7.48VLWQYY136 pKa = 11.33IYY138 pKa = 10.63FQTEE142 pKa = 3.35SDD144 pKa = 3.33TWSKK148 pKa = 10.13TEE150 pKa = 4.0GKK152 pKa = 10.15LDD154 pKa = 3.78NLGLYY159 pKa = 10.27YY160 pKa = 10.47LDD162 pKa = 3.32EE163 pKa = 4.41HH164 pKa = 6.48HH165 pKa = 7.05FKK167 pKa = 10.86VYY169 pKa = 10.68YY170 pKa = 9.83VDD172 pKa = 4.31FRR174 pKa = 11.84KK175 pKa = 9.92EE176 pKa = 3.6AEE178 pKa = 4.46KK179 pKa = 10.73YY180 pKa = 10.05SKK182 pKa = 9.16TGRR185 pKa = 11.84YY186 pKa = 7.89EE187 pKa = 4.04VLSKK191 pKa = 10.25LTTPAVPTSPFAATGPATGGDD212 pKa = 3.66ASTSGATARR221 pKa = 11.84PSTPKK226 pKa = 10.4KK227 pKa = 9.82KK228 pKa = 10.35ALPTTPRR235 pKa = 11.84RR236 pKa = 11.84SRR238 pKa = 11.84RR239 pKa = 11.84YY240 pKa = 8.86LRR242 pKa = 11.84PSSRR246 pKa = 11.84EE247 pKa = 3.57TPRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84GFGRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84EE259 pKa = 4.23GEE261 pKa = 4.12LAGHH265 pKa = 5.87STPSPGLVPPSPGEE279 pKa = 3.76VGQRR283 pKa = 11.84TQTTPKK289 pKa = 10.5GNRR292 pKa = 11.84TRR294 pKa = 11.84LARR297 pKa = 11.84LLLDD301 pKa = 4.38AKK303 pKa = 10.24DD304 pKa = 3.72PPVIILKK311 pKa = 10.32GGPNSLKK318 pKa = 10.35CIRR321 pKa = 11.84YY322 pKa = 8.33RR323 pKa = 11.84LKK325 pKa = 11.06SKK327 pKa = 10.52FSSLFSRR334 pKa = 11.84ASTTWQWTVSDD345 pKa = 3.73GTEE348 pKa = 3.14RR349 pKa = 11.84WGRR352 pKa = 11.84SRR354 pKa = 11.84MLLSFKK360 pKa = 10.07DD361 pKa = 3.42TAQRR365 pKa = 11.84EE366 pKa = 4.62LFLHH370 pKa = 6.0TVQLPKK376 pKa = 10.37SVQYY380 pKa = 11.24SLGSFDD386 pKa = 5.74DD387 pKa = 3.99LL388 pKa = 5.16
MM1 pKa = 7.63EE2 pKa = 5.32NLSKK6 pKa = 10.98ALDD9 pKa = 3.73SLQEE13 pKa = 4.1EE14 pKa = 4.64LLSLYY19 pKa = 9.66EE20 pKa = 4.39KK21 pKa = 10.91NSGDD25 pKa = 3.11IADD28 pKa = 5.35QIRR31 pKa = 11.84HH32 pKa = 4.87WNLVRR37 pKa = 11.84KK38 pKa = 8.51EE39 pKa = 3.68QVLFHH44 pKa = 6.48FARR47 pKa = 11.84KK48 pKa = 9.78NGVSRR53 pKa = 11.84IGMSPVPPQSVSQQKK68 pKa = 10.08AKK70 pKa = 10.69SAIEE74 pKa = 3.9QEE76 pKa = 4.17ILLQSLADD84 pKa = 3.56SQYY87 pKa = 11.0GSEE90 pKa = 4.28RR91 pKa = 11.84WTLSDD96 pKa = 3.23TSKK99 pKa = 10.92EE100 pKa = 3.9RR101 pKa = 11.84LLADD105 pKa = 3.91PPYY108 pKa = 10.54CFKK111 pKa = 11.33KK112 pKa = 10.39NGKK115 pKa = 8.86QVDD118 pKa = 3.59VRR120 pKa = 11.84FDD122 pKa = 3.63EE123 pKa = 4.71QPDD126 pKa = 3.49NLVRR130 pKa = 11.84YY131 pKa = 7.48VLWQYY136 pKa = 11.33IYY138 pKa = 10.63FQTEE142 pKa = 3.35SDD144 pKa = 3.33TWSKK148 pKa = 10.13TEE150 pKa = 4.0GKK152 pKa = 10.15LDD154 pKa = 3.78NLGLYY159 pKa = 10.27YY160 pKa = 10.47LDD162 pKa = 3.32EE163 pKa = 4.41HH164 pKa = 6.48HH165 pKa = 7.05FKK167 pKa = 10.86VYY169 pKa = 10.68YY170 pKa = 9.83VDD172 pKa = 4.31FRR174 pKa = 11.84KK175 pKa = 9.92EE176 pKa = 3.6AEE178 pKa = 4.46KK179 pKa = 10.73YY180 pKa = 10.05SKK182 pKa = 9.16TGRR185 pKa = 11.84YY186 pKa = 7.89EE187 pKa = 4.04VLSKK191 pKa = 10.25LTTPAVPTSPFAATGPATGGDD212 pKa = 3.66ASTSGATARR221 pKa = 11.84PSTPKK226 pKa = 10.4KK227 pKa = 9.82KK228 pKa = 10.35ALPTTPRR235 pKa = 11.84RR236 pKa = 11.84SRR238 pKa = 11.84RR239 pKa = 11.84YY240 pKa = 8.86LRR242 pKa = 11.84PSSRR246 pKa = 11.84EE247 pKa = 3.57TPRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84GFGRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84EE259 pKa = 4.23GEE261 pKa = 4.12LAGHH265 pKa = 5.87STPSPGLVPPSPGEE279 pKa = 3.76VGQRR283 pKa = 11.84TQTTPKK289 pKa = 10.5GNRR292 pKa = 11.84TRR294 pKa = 11.84LARR297 pKa = 11.84LLLDD301 pKa = 4.38AKK303 pKa = 10.24DD304 pKa = 3.72PPVIILKK311 pKa = 10.32GGPNSLKK318 pKa = 10.35CIRR321 pKa = 11.84YY322 pKa = 8.33RR323 pKa = 11.84LKK325 pKa = 11.06SKK327 pKa = 10.52FSSLFSRR334 pKa = 11.84ASTTWQWTVSDD345 pKa = 3.73GTEE348 pKa = 3.14RR349 pKa = 11.84WGRR352 pKa = 11.84SRR354 pKa = 11.84MLLSFKK360 pKa = 10.07DD361 pKa = 3.42TAQRR365 pKa = 11.84EE366 pKa = 4.62LFLHH370 pKa = 6.0TVQLPKK376 pKa = 10.37SVQYY380 pKa = 11.24SLGSFDD386 pKa = 5.74DD387 pKa = 3.99LL388 pKa = 5.16
Molecular weight: 44.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2355 |
97 |
603 |
336.4 |
38.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.308 ± 0.304 | 1.953 ± 0.63 |
6.709 ± 0.548 | 6.709 ± 0.61 |
4.798 ± 0.504 | 5.605 ± 0.437 |
1.529 ± 0.131 | 4.756 ± 0.859 |
5.86 ± 0.764 | 9.894 ± 0.744 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.104 ± 0.267 | 3.355 ± 0.638 |
6.582 ± 1.231 | 4.798 ± 0.331 |
6.582 ± 0.838 | 7.856 ± 0.877 |
6.285 ± 0.787 | 5.478 ± 0.433 |
1.359 ± 0.337 | 3.482 ± 0.248 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
