
Rhodohalobacter barkolensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Rhodohalobacter
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
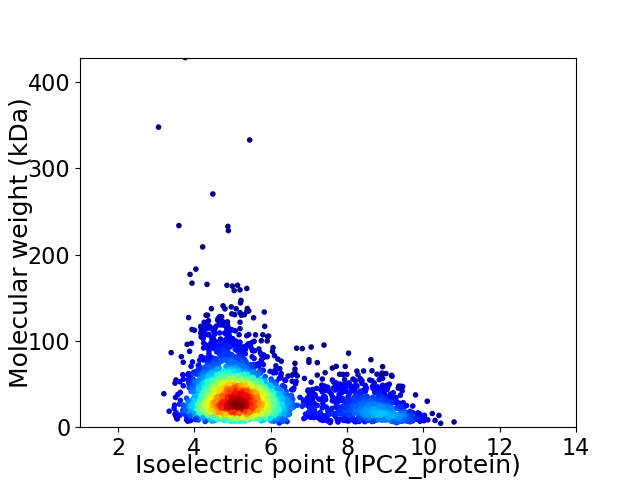
Virtual 2D-PAGE plot for 3017 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N0VFE7|A0A2N0VFE7_9BACT Uncharacterized protein OS=Rhodohalobacter barkolensis OX=2053187 GN=CWD77_12750 PE=4 SV=1
MM1 pKa = 7.5NNLKK5 pKa = 10.66VNFMKK10 pKa = 10.62PIKK13 pKa = 10.1RR14 pKa = 11.84LMLVTLIGVSFMSCDD29 pKa = 4.29DD30 pKa = 4.41FGDD33 pKa = 5.06LNDD36 pKa = 4.97DD37 pKa = 4.19PNNPSQVSPDD47 pKa = 3.44QLLTDD52 pKa = 3.91SQRR55 pKa = 11.84NMSDD59 pKa = 2.97VIGAVNGTLYY69 pKa = 10.16VQYY72 pKa = 10.34IAEE75 pKa = 4.22TQYY78 pKa = 11.41TDD80 pKa = 3.06AQEE83 pKa = 4.07YY84 pKa = 7.17RR85 pKa = 11.84TTNASFYY92 pKa = 10.52FWYY95 pKa = 9.56TDD97 pKa = 4.61PIQNLQTIIDD107 pKa = 4.51LNTDD111 pKa = 3.19EE112 pKa = 4.58EE113 pKa = 4.65TRR115 pKa = 11.84DD116 pKa = 3.37QASALGSNANQIAVARR132 pKa = 11.84ILKK135 pKa = 10.16AYY137 pKa = 8.87FYY139 pKa = 11.42QMMTDD144 pKa = 2.96RR145 pKa = 11.84WGMIPYY151 pKa = 9.82SEE153 pKa = 4.37ALQGEE158 pKa = 4.84EE159 pKa = 4.69DD160 pKa = 4.37FSPAYY165 pKa = 10.12DD166 pKa = 3.71SQEE169 pKa = 4.45DD170 pKa = 3.99IYY172 pKa = 11.22EE173 pKa = 4.42AIITEE178 pKa = 4.33LKK180 pKa = 10.14EE181 pKa = 3.84AADD184 pKa = 3.78QIEE187 pKa = 4.33LSEE190 pKa = 4.8AGVSGDD196 pKa = 3.47ILFSGDD202 pKa = 3.04MEE204 pKa = 4.32QWVLFANSLRR214 pKa = 11.84ARR216 pKa = 11.84AALRR220 pKa = 11.84IADD223 pKa = 3.63VGGDD227 pKa = 3.59VSVDD231 pKa = 3.49PAAEE235 pKa = 3.85FADD238 pKa = 4.01AVDD241 pKa = 4.99DD242 pKa = 4.22GLINSDD248 pKa = 3.32VLYY251 pKa = 9.38PYY253 pKa = 10.4QSNADD258 pKa = 4.74DD259 pKa = 3.98EE260 pKa = 4.85NPWYY264 pKa = 10.91SRR266 pKa = 11.84FQTRR270 pKa = 11.84TDD272 pKa = 3.5YY273 pKa = 11.34AISEE277 pKa = 4.53TIADD281 pKa = 3.85YY282 pKa = 10.03MKK284 pKa = 10.78GLEE287 pKa = 4.2DD288 pKa = 5.45DD289 pKa = 5.07RR290 pKa = 11.84ILVYY294 pKa = 11.0ANPAPNYY301 pKa = 10.11SNDD304 pKa = 3.74DD305 pKa = 3.55GVVTFDD311 pKa = 5.03EE312 pKa = 4.25IRR314 pKa = 11.84GMPFLEE320 pKa = 5.0DD321 pKa = 3.36AGEE324 pKa = 4.17LEE326 pKa = 4.12NAEE329 pKa = 3.91ISFPGSAIGAGGPGVGDD346 pKa = 3.25QSAPLPIITVAEE358 pKa = 3.92MHH360 pKa = 6.38FAMAEE365 pKa = 3.78AVEE368 pKa = 4.56RR369 pKa = 11.84GWITGAAADD378 pKa = 4.1YY379 pKa = 10.44YY380 pKa = 11.02YY381 pKa = 11.08DD382 pKa = 4.4GIEE385 pKa = 4.51ASWEE389 pKa = 3.66QWGVYY394 pKa = 10.33DD395 pKa = 4.25DD396 pKa = 4.73TNFADD401 pKa = 5.14YY402 pKa = 10.48IAQTEE407 pKa = 4.42VAYY410 pKa = 8.71GTDD413 pKa = 3.42DD414 pKa = 3.36WDD416 pKa = 3.96VQIGTQKK423 pKa = 10.2WIALFPHH430 pKa = 7.25GYY432 pKa = 9.61EE433 pKa = 4.86GWAEE437 pKa = 3.96WRR439 pKa = 11.84RR440 pKa = 11.84LGQPEE445 pKa = 4.35LTPNQFGVGTDD456 pKa = 3.3PQIPVRR462 pKa = 11.84FTYY465 pKa = 10.07PSSEE469 pKa = 3.72NTLNQEE475 pKa = 3.95NYY477 pKa = 8.51EE478 pKa = 4.22AAVQAQGPDD487 pKa = 3.48SPNTRR492 pKa = 11.84IWWDD496 pKa = 2.84VDD498 pKa = 3.13
MM1 pKa = 7.5NNLKK5 pKa = 10.66VNFMKK10 pKa = 10.62PIKK13 pKa = 10.1RR14 pKa = 11.84LMLVTLIGVSFMSCDD29 pKa = 4.29DD30 pKa = 4.41FGDD33 pKa = 5.06LNDD36 pKa = 4.97DD37 pKa = 4.19PNNPSQVSPDD47 pKa = 3.44QLLTDD52 pKa = 3.91SQRR55 pKa = 11.84NMSDD59 pKa = 2.97VIGAVNGTLYY69 pKa = 10.16VQYY72 pKa = 10.34IAEE75 pKa = 4.22TQYY78 pKa = 11.41TDD80 pKa = 3.06AQEE83 pKa = 4.07YY84 pKa = 7.17RR85 pKa = 11.84TTNASFYY92 pKa = 10.52FWYY95 pKa = 9.56TDD97 pKa = 4.61PIQNLQTIIDD107 pKa = 4.51LNTDD111 pKa = 3.19EE112 pKa = 4.58EE113 pKa = 4.65TRR115 pKa = 11.84DD116 pKa = 3.37QASALGSNANQIAVARR132 pKa = 11.84ILKK135 pKa = 10.16AYY137 pKa = 8.87FYY139 pKa = 11.42QMMTDD144 pKa = 2.96RR145 pKa = 11.84WGMIPYY151 pKa = 9.82SEE153 pKa = 4.37ALQGEE158 pKa = 4.84EE159 pKa = 4.69DD160 pKa = 4.37FSPAYY165 pKa = 10.12DD166 pKa = 3.71SQEE169 pKa = 4.45DD170 pKa = 3.99IYY172 pKa = 11.22EE173 pKa = 4.42AIITEE178 pKa = 4.33LKK180 pKa = 10.14EE181 pKa = 3.84AADD184 pKa = 3.78QIEE187 pKa = 4.33LSEE190 pKa = 4.8AGVSGDD196 pKa = 3.47ILFSGDD202 pKa = 3.04MEE204 pKa = 4.32QWVLFANSLRR214 pKa = 11.84ARR216 pKa = 11.84AALRR220 pKa = 11.84IADD223 pKa = 3.63VGGDD227 pKa = 3.59VSVDD231 pKa = 3.49PAAEE235 pKa = 3.85FADD238 pKa = 4.01AVDD241 pKa = 4.99DD242 pKa = 4.22GLINSDD248 pKa = 3.32VLYY251 pKa = 9.38PYY253 pKa = 10.4QSNADD258 pKa = 4.74DD259 pKa = 3.98EE260 pKa = 4.85NPWYY264 pKa = 10.91SRR266 pKa = 11.84FQTRR270 pKa = 11.84TDD272 pKa = 3.5YY273 pKa = 11.34AISEE277 pKa = 4.53TIADD281 pKa = 3.85YY282 pKa = 10.03MKK284 pKa = 10.78GLEE287 pKa = 4.2DD288 pKa = 5.45DD289 pKa = 5.07RR290 pKa = 11.84ILVYY294 pKa = 11.0ANPAPNYY301 pKa = 10.11SNDD304 pKa = 3.74DD305 pKa = 3.55GVVTFDD311 pKa = 5.03EE312 pKa = 4.25IRR314 pKa = 11.84GMPFLEE320 pKa = 5.0DD321 pKa = 3.36AGEE324 pKa = 4.17LEE326 pKa = 4.12NAEE329 pKa = 3.91ISFPGSAIGAGGPGVGDD346 pKa = 3.25QSAPLPIITVAEE358 pKa = 3.92MHH360 pKa = 6.38FAMAEE365 pKa = 3.78AVEE368 pKa = 4.56RR369 pKa = 11.84GWITGAAADD378 pKa = 4.1YY379 pKa = 10.44YY380 pKa = 11.02YY381 pKa = 11.08DD382 pKa = 4.4GIEE385 pKa = 4.51ASWEE389 pKa = 3.66QWGVYY394 pKa = 10.33DD395 pKa = 4.25DD396 pKa = 4.73TNFADD401 pKa = 5.14YY402 pKa = 10.48IAQTEE407 pKa = 4.42VAYY410 pKa = 8.71GTDD413 pKa = 3.42DD414 pKa = 3.36WDD416 pKa = 3.96VQIGTQKK423 pKa = 10.2WIALFPHH430 pKa = 7.25GYY432 pKa = 9.61EE433 pKa = 4.86GWAEE437 pKa = 3.96WRR439 pKa = 11.84RR440 pKa = 11.84LGQPEE445 pKa = 4.35LTPNQFGVGTDD456 pKa = 3.3PQIPVRR462 pKa = 11.84FTYY465 pKa = 10.07PSSEE469 pKa = 3.72NTLNQEE475 pKa = 3.95NYY477 pKa = 8.51EE478 pKa = 4.22AAVQAQGPDD487 pKa = 3.48SPNTRR492 pKa = 11.84IWWDD496 pKa = 2.84VDD498 pKa = 3.13
Molecular weight: 55.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N0VKN7|A0A2N0VKN7_9BACT Ribose 5-phosphate isomerase B OS=Rhodohalobacter barkolensis OX=2053187 GN=rpiB PE=3 SV=1
MM1 pKa = 7.57AHH3 pKa = 7.61PKK5 pKa = 10.32RR6 pKa = 11.84KK7 pKa = 7.65TSKK10 pKa = 8.0TRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.28KK15 pKa = 10.96RR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.37HH20 pKa = 6.4RR21 pKa = 11.84VAEE24 pKa = 4.22IPLSEE29 pKa = 4.28CTNCGAMHH37 pKa = 7.43RR38 pKa = 11.84YY39 pKa = 8.46HH40 pKa = 6.61HH41 pKa = 6.56VCMEE45 pKa = 4.23CGYY48 pKa = 10.15YY49 pKa = 10.05RR50 pKa = 11.84GRR52 pKa = 11.84QVLNVSKK59 pKa = 11.04
MM1 pKa = 7.57AHH3 pKa = 7.61PKK5 pKa = 10.32RR6 pKa = 11.84KK7 pKa = 7.65TSKK10 pKa = 8.0TRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.28KK15 pKa = 10.96RR16 pKa = 11.84RR17 pKa = 11.84SHH19 pKa = 6.37HH20 pKa = 6.4RR21 pKa = 11.84VAEE24 pKa = 4.22IPLSEE29 pKa = 4.28CTNCGAMHH37 pKa = 7.43RR38 pKa = 11.84YY39 pKa = 8.46HH40 pKa = 6.61HH41 pKa = 6.56VCMEE45 pKa = 4.23CGYY48 pKa = 10.15YY49 pKa = 10.05RR50 pKa = 11.84GRR52 pKa = 11.84QVLNVSKK59 pKa = 11.04
Molecular weight: 7.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1043729 |
38 |
3927 |
345.9 |
38.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.565 ± 0.038 | 0.576 ± 0.014 |
6.231 ± 0.044 | 7.725 ± 0.047 |
4.813 ± 0.037 | 6.92 ± 0.046 |
1.921 ± 0.025 | 7.205 ± 0.037 |
5.586 ± 0.061 | 9.301 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.485 ± 0.022 | 4.919 ± 0.033 |
3.898 ± 0.024 | 3.651 ± 0.027 |
4.683 ± 0.035 | 6.93 ± 0.045 |
5.372 ± 0.037 | 6.452 ± 0.038 |
1.199 ± 0.021 | 3.568 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
