
Tomato aspermy virus (TAV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Cucumovirus
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
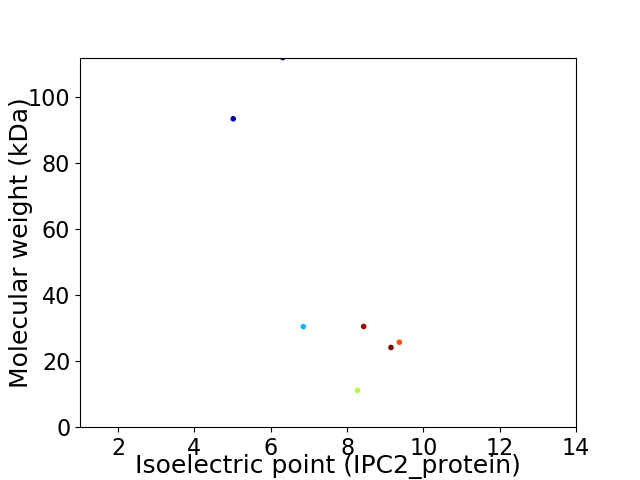
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q88619|Q88619_TAV Capsid protein OS=Tomato aspermy virus OX=12315 GN=cp PE=2 SV=1
MM1 pKa = 7.19STFSLEE7 pKa = 3.74NCLNGSYY14 pKa = 10.84GVDD17 pKa = 3.2TPEE20 pKa = 3.55EE21 pKa = 3.98VTFIRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84DD29 pKa = 3.63ANPSAFHH36 pKa = 6.99GEE38 pKa = 3.94EE39 pKa = 4.11TVPSNTTEE47 pKa = 3.87VHH49 pKa = 5.87SEE51 pKa = 4.17SVSYY55 pKa = 11.13SSDD58 pKa = 3.46FEE60 pKa = 5.35GYY62 pKa = 10.22SCDD65 pKa = 4.68SDD67 pKa = 3.63FTGDD71 pKa = 3.41NQYY74 pKa = 11.51SSFDD78 pKa = 3.65GADD81 pKa = 3.71SLLLSYY87 pKa = 10.79DD88 pKa = 2.79IDD90 pKa = 3.92EE91 pKa = 5.26YY92 pKa = 11.13IDD94 pKa = 3.86LSEE97 pKa = 4.89SEE99 pKa = 4.47LRR101 pKa = 11.84VLFDD105 pKa = 4.15EE106 pKa = 4.8MVQPIHH112 pKa = 6.84FGMVLSPTFSRR123 pKa = 11.84NRR125 pKa = 11.84FLSCLSLAKK134 pKa = 10.16SVVVAPLYY142 pKa = 10.84DD143 pKa = 3.45PEE145 pKa = 4.22RR146 pKa = 11.84TLRR149 pKa = 11.84PFPEE153 pKa = 4.24LVKK156 pKa = 10.6NLYY159 pKa = 10.62SGVHH163 pKa = 7.03DD164 pKa = 4.7IYY166 pKa = 11.3LSDD169 pKa = 3.92SEE171 pKa = 4.31EE172 pKa = 3.92AAISSVTDD180 pKa = 3.79TIEE183 pKa = 4.74GYY185 pKa = 8.49TFSPRR190 pKa = 11.84DD191 pKa = 3.3VVDD194 pKa = 3.72NYY196 pKa = 10.63EE197 pKa = 4.4PPPLCSVCGLIAYY210 pKa = 7.44QCPHH214 pKa = 6.94YY215 pKa = 10.52DD216 pKa = 3.24INSLRR221 pKa = 11.84KK222 pKa = 8.83NCAEE226 pKa = 3.69MTIAHH231 pKa = 7.57DD232 pKa = 3.93YY233 pKa = 10.26PIEE236 pKa = 4.22GLCGLIDD243 pKa = 5.32DD244 pKa = 4.6ATLLKK249 pKa = 10.91NLGSFLLPIQCEE261 pKa = 4.31YY262 pKa = 10.94TKK264 pKa = 10.9LPEE267 pKa = 4.32PTLITPPEE275 pKa = 4.12LTRR278 pKa = 11.84PTDD281 pKa = 3.25RR282 pKa = 11.84VTVDD286 pKa = 4.22LLQAICDD293 pKa = 3.95STLPTHH299 pKa = 6.17VCYY302 pKa = 10.56DD303 pKa = 3.57DD304 pKa = 5.03TYY306 pKa = 10.44HH307 pKa = 5.85QTFIEE312 pKa = 4.24NADD315 pKa = 3.46YY316 pKa = 11.16SVDD319 pKa = 3.3IDD321 pKa = 4.06RR322 pKa = 11.84VRR324 pKa = 11.84LKK326 pKa = 10.83QSDD329 pKa = 4.17LLAKK333 pKa = 10.62VIDD336 pKa = 4.21EE337 pKa = 4.33GHH339 pKa = 7.07LKK341 pKa = 10.23PVLNTGSGQKK351 pKa = 10.37RR352 pKa = 11.84IGTTRR357 pKa = 11.84EE358 pKa = 3.64VLCAIKK364 pKa = 10.17KK365 pKa = 10.03RR366 pKa = 11.84NADD369 pKa = 3.51VPEE372 pKa = 4.28LCGSVNLKK380 pKa = 10.34RR381 pKa = 11.84LSDD384 pKa = 3.91DD385 pKa = 3.37VAEE388 pKa = 4.51CFMLSFMNGDD398 pKa = 4.12KK399 pKa = 10.85LCSSNFINIVSDD411 pKa = 3.79FHH413 pKa = 9.25AYY415 pKa = 7.45MSKK418 pKa = 8.18WQSVLSYY425 pKa = 11.52DD426 pKa = 5.54DD427 pKa = 5.9LPDD430 pKa = 4.0LNAEE434 pKa = 3.83NLQFYY439 pKa = 9.9EE440 pKa = 4.57HH441 pKa = 6.27MVKK444 pKa = 10.41SDD446 pKa = 3.47VKK448 pKa = 10.83PSVTDD453 pKa = 3.07TLNIDD458 pKa = 3.66RR459 pKa = 11.84PLPATITFHH468 pKa = 7.31RR469 pKa = 11.84KK470 pKa = 8.36QLTSQFSPLFTALFQRR486 pKa = 11.84FQRR489 pKa = 11.84CLTKK493 pKa = 10.46RR494 pKa = 11.84VILPVGKK501 pKa = 10.07ISSLEE506 pKa = 3.83IKK508 pKa = 10.53DD509 pKa = 3.84FSVLNKK515 pKa = 9.82FCLEE519 pKa = 3.67IDD521 pKa = 3.44LSKK524 pKa = 10.66FDD526 pKa = 3.77KK527 pKa = 11.17SQGEE531 pKa = 4.26LHH533 pKa = 7.14LMIQEE538 pKa = 5.14GILNRR543 pKa = 11.84LGCPVHH549 pKa = 7.19ISKK552 pKa = 8.59WWCDD556 pKa = 4.24FHH558 pKa = 8.96RR559 pKa = 11.84MSYY562 pKa = 10.37IKK564 pKa = 10.2DD565 pKa = 3.1KK566 pKa = 10.84RR567 pKa = 11.84AGVSMPISFQRR578 pKa = 11.84RR579 pKa = 11.84TGDD582 pKa = 2.78AFTYY586 pKa = 10.33FGNTLVTMSMFAWCYY601 pKa = 8.26DD602 pKa = 3.0TSQFDD607 pKa = 3.68KK608 pKa = 11.33LIFSGDD614 pKa = 3.51DD615 pKa = 3.19SLGFSIKK622 pKa = 10.7APVGDD627 pKa = 4.02PSLFTSLFNMEE638 pKa = 4.61AKK640 pKa = 10.52VMEE643 pKa = 4.39PSVPYY648 pKa = 10.05ICSKK652 pKa = 10.76FLLTDD657 pKa = 3.47DD658 pKa = 4.79FGNTFSVPDD667 pKa = 4.45PLRR670 pKa = 11.84EE671 pKa = 3.88IQRR674 pKa = 11.84LGSKK678 pKa = 10.16KK679 pKa = 10.13IPLDD683 pKa = 3.6EE684 pKa = 5.73DD685 pKa = 3.72DD686 pKa = 5.28RR687 pKa = 11.84SLHH690 pKa = 5.02AHH692 pKa = 6.06FMSFVDD698 pKa = 3.91RR699 pKa = 11.84LKK701 pKa = 10.95FLNHH705 pKa = 6.01MNQTSMTQLSLFYY718 pKa = 10.42EE719 pKa = 4.17MKK721 pKa = 10.12YY722 pKa = 10.38RR723 pKa = 11.84KK724 pKa = 10.14SGDD727 pKa = 4.06DD728 pKa = 3.06ILLVLGAFNKK738 pKa = 8.67YY739 pKa = 6.24TANFNAYY746 pKa = 9.22KK747 pKa = 10.1EE748 pKa = 4.42LYY750 pKa = 10.0YY751 pKa = 10.4SEE753 pKa = 4.76KK754 pKa = 9.84QQCALINSFSISDD767 pKa = 4.44LIVGRR772 pKa = 11.84GKK774 pKa = 10.36SSKK777 pKa = 9.99ASRR780 pKa = 11.84RR781 pKa = 11.84KK782 pKa = 9.69AVEE785 pKa = 4.0SNGKK789 pKa = 8.84HH790 pKa = 6.11RR791 pKa = 11.84DD792 pKa = 3.1PSTRR796 pKa = 11.84DD797 pKa = 3.17HH798 pKa = 6.92SKK800 pKa = 10.99VGTDD804 pKa = 3.38EE805 pKa = 4.46SKK807 pKa = 9.48EE808 pKa = 4.23TSTEE812 pKa = 3.76EE813 pKa = 3.99TTQTEE818 pKa = 4.23PQGAGSQKK826 pKa = 10.86SKK828 pKa = 11.24
MM1 pKa = 7.19STFSLEE7 pKa = 3.74NCLNGSYY14 pKa = 10.84GVDD17 pKa = 3.2TPEE20 pKa = 3.55EE21 pKa = 3.98VTFIRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84DD29 pKa = 3.63ANPSAFHH36 pKa = 6.99GEE38 pKa = 3.94EE39 pKa = 4.11TVPSNTTEE47 pKa = 3.87VHH49 pKa = 5.87SEE51 pKa = 4.17SVSYY55 pKa = 11.13SSDD58 pKa = 3.46FEE60 pKa = 5.35GYY62 pKa = 10.22SCDD65 pKa = 4.68SDD67 pKa = 3.63FTGDD71 pKa = 3.41NQYY74 pKa = 11.51SSFDD78 pKa = 3.65GADD81 pKa = 3.71SLLLSYY87 pKa = 10.79DD88 pKa = 2.79IDD90 pKa = 3.92EE91 pKa = 5.26YY92 pKa = 11.13IDD94 pKa = 3.86LSEE97 pKa = 4.89SEE99 pKa = 4.47LRR101 pKa = 11.84VLFDD105 pKa = 4.15EE106 pKa = 4.8MVQPIHH112 pKa = 6.84FGMVLSPTFSRR123 pKa = 11.84NRR125 pKa = 11.84FLSCLSLAKK134 pKa = 10.16SVVVAPLYY142 pKa = 10.84DD143 pKa = 3.45PEE145 pKa = 4.22RR146 pKa = 11.84TLRR149 pKa = 11.84PFPEE153 pKa = 4.24LVKK156 pKa = 10.6NLYY159 pKa = 10.62SGVHH163 pKa = 7.03DD164 pKa = 4.7IYY166 pKa = 11.3LSDD169 pKa = 3.92SEE171 pKa = 4.31EE172 pKa = 3.92AAISSVTDD180 pKa = 3.79TIEE183 pKa = 4.74GYY185 pKa = 8.49TFSPRR190 pKa = 11.84DD191 pKa = 3.3VVDD194 pKa = 3.72NYY196 pKa = 10.63EE197 pKa = 4.4PPPLCSVCGLIAYY210 pKa = 7.44QCPHH214 pKa = 6.94YY215 pKa = 10.52DD216 pKa = 3.24INSLRR221 pKa = 11.84KK222 pKa = 8.83NCAEE226 pKa = 3.69MTIAHH231 pKa = 7.57DD232 pKa = 3.93YY233 pKa = 10.26PIEE236 pKa = 4.22GLCGLIDD243 pKa = 5.32DD244 pKa = 4.6ATLLKK249 pKa = 10.91NLGSFLLPIQCEE261 pKa = 4.31YY262 pKa = 10.94TKK264 pKa = 10.9LPEE267 pKa = 4.32PTLITPPEE275 pKa = 4.12LTRR278 pKa = 11.84PTDD281 pKa = 3.25RR282 pKa = 11.84VTVDD286 pKa = 4.22LLQAICDD293 pKa = 3.95STLPTHH299 pKa = 6.17VCYY302 pKa = 10.56DD303 pKa = 3.57DD304 pKa = 5.03TYY306 pKa = 10.44HH307 pKa = 5.85QTFIEE312 pKa = 4.24NADD315 pKa = 3.46YY316 pKa = 11.16SVDD319 pKa = 3.3IDD321 pKa = 4.06RR322 pKa = 11.84VRR324 pKa = 11.84LKK326 pKa = 10.83QSDD329 pKa = 4.17LLAKK333 pKa = 10.62VIDD336 pKa = 4.21EE337 pKa = 4.33GHH339 pKa = 7.07LKK341 pKa = 10.23PVLNTGSGQKK351 pKa = 10.37RR352 pKa = 11.84IGTTRR357 pKa = 11.84EE358 pKa = 3.64VLCAIKK364 pKa = 10.17KK365 pKa = 10.03RR366 pKa = 11.84NADD369 pKa = 3.51VPEE372 pKa = 4.28LCGSVNLKK380 pKa = 10.34RR381 pKa = 11.84LSDD384 pKa = 3.91DD385 pKa = 3.37VAEE388 pKa = 4.51CFMLSFMNGDD398 pKa = 4.12KK399 pKa = 10.85LCSSNFINIVSDD411 pKa = 3.79FHH413 pKa = 9.25AYY415 pKa = 7.45MSKK418 pKa = 8.18WQSVLSYY425 pKa = 11.52DD426 pKa = 5.54DD427 pKa = 5.9LPDD430 pKa = 4.0LNAEE434 pKa = 3.83NLQFYY439 pKa = 9.9EE440 pKa = 4.57HH441 pKa = 6.27MVKK444 pKa = 10.41SDD446 pKa = 3.47VKK448 pKa = 10.83PSVTDD453 pKa = 3.07TLNIDD458 pKa = 3.66RR459 pKa = 11.84PLPATITFHH468 pKa = 7.31RR469 pKa = 11.84KK470 pKa = 8.36QLTSQFSPLFTALFQRR486 pKa = 11.84FQRR489 pKa = 11.84CLTKK493 pKa = 10.46RR494 pKa = 11.84VILPVGKK501 pKa = 10.07ISSLEE506 pKa = 3.83IKK508 pKa = 10.53DD509 pKa = 3.84FSVLNKK515 pKa = 9.82FCLEE519 pKa = 3.67IDD521 pKa = 3.44LSKK524 pKa = 10.66FDD526 pKa = 3.77KK527 pKa = 11.17SQGEE531 pKa = 4.26LHH533 pKa = 7.14LMIQEE538 pKa = 5.14GILNRR543 pKa = 11.84LGCPVHH549 pKa = 7.19ISKK552 pKa = 8.59WWCDD556 pKa = 4.24FHH558 pKa = 8.96RR559 pKa = 11.84MSYY562 pKa = 10.37IKK564 pKa = 10.2DD565 pKa = 3.1KK566 pKa = 10.84RR567 pKa = 11.84AGVSMPISFQRR578 pKa = 11.84RR579 pKa = 11.84TGDD582 pKa = 2.78AFTYY586 pKa = 10.33FGNTLVTMSMFAWCYY601 pKa = 8.26DD602 pKa = 3.0TSQFDD607 pKa = 3.68KK608 pKa = 11.33LIFSGDD614 pKa = 3.51DD615 pKa = 3.19SLGFSIKK622 pKa = 10.7APVGDD627 pKa = 4.02PSLFTSLFNMEE638 pKa = 4.61AKK640 pKa = 10.52VMEE643 pKa = 4.39PSVPYY648 pKa = 10.05ICSKK652 pKa = 10.76FLLTDD657 pKa = 3.47DD658 pKa = 4.79FGNTFSVPDD667 pKa = 4.45PLRR670 pKa = 11.84EE671 pKa = 3.88IQRR674 pKa = 11.84LGSKK678 pKa = 10.16KK679 pKa = 10.13IPLDD683 pKa = 3.6EE684 pKa = 5.73DD685 pKa = 3.72DD686 pKa = 5.28RR687 pKa = 11.84SLHH690 pKa = 5.02AHH692 pKa = 6.06FMSFVDD698 pKa = 3.91RR699 pKa = 11.84LKK701 pKa = 10.95FLNHH705 pKa = 6.01MNQTSMTQLSLFYY718 pKa = 10.42EE719 pKa = 4.17MKK721 pKa = 10.12YY722 pKa = 10.38RR723 pKa = 11.84KK724 pKa = 10.14SGDD727 pKa = 4.06DD728 pKa = 3.06ILLVLGAFNKK738 pKa = 8.67YY739 pKa = 6.24TANFNAYY746 pKa = 9.22KK747 pKa = 10.1EE748 pKa = 4.42LYY750 pKa = 10.0YY751 pKa = 10.4SEE753 pKa = 4.76KK754 pKa = 9.84QQCALINSFSISDD767 pKa = 4.44LIVGRR772 pKa = 11.84GKK774 pKa = 10.36SSKK777 pKa = 9.99ASRR780 pKa = 11.84RR781 pKa = 11.84KK782 pKa = 9.69AVEE785 pKa = 4.0SNGKK789 pKa = 8.84HH790 pKa = 6.11RR791 pKa = 11.84DD792 pKa = 3.1PSTRR796 pKa = 11.84DD797 pKa = 3.17HH798 pKa = 6.92SKK800 pKa = 10.99VGTDD804 pKa = 3.38EE805 pKa = 4.46SKK807 pKa = 9.48EE808 pKa = 4.23TSTEE812 pKa = 3.76EE813 pKa = 3.99TTQTEE818 pKa = 4.23PQGAGSQKK826 pKa = 10.86SKK828 pKa = 11.24
Molecular weight: 93.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P28931|1A_TAV Replication protein 1a OS=Tomato aspermy virus OX=12315 GN=ORF1a PE=3 SV=1
MM1 pKa = 7.26AQNGTGGGSRR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84NNNNNNSTARR28 pKa = 11.84DD29 pKa = 3.58KK30 pKa = 11.54ALLALTQQVNRR41 pKa = 11.84LANIASSSAPSLQHH55 pKa = 5.37PTFIASKK62 pKa = 9.33KK63 pKa = 9.13CRR65 pKa = 11.84AGYY68 pKa = 8.61TYY70 pKa = 10.49TSLDD74 pKa = 3.31VRR76 pKa = 11.84PTRR79 pKa = 11.84TEE81 pKa = 3.58KK82 pKa = 10.9DD83 pKa = 2.95KK84 pKa = 11.8SFGQRR89 pKa = 11.84LIIPVPVSEE98 pKa = 4.23YY99 pKa = 9.43PKK101 pKa = 10.66KK102 pKa = 10.4KK103 pKa = 9.72VSCVQVRR110 pKa = 11.84LNPSPKK116 pKa = 9.47FNSTIWVSLRR126 pKa = 11.84RR127 pKa = 11.84LDD129 pKa = 3.62EE130 pKa = 4.33TTLLTSEE137 pKa = 4.43NVFKK141 pKa = 11.33LFTDD145 pKa = 4.24GLAAVLIYY153 pKa = 9.4QHH155 pKa = 6.54VPTGIQPNNKK165 pKa = 8.16ITFDD169 pKa = 3.57MSNVGAEE176 pKa = 3.78IGDD179 pKa = 3.54MGKK182 pKa = 10.16YY183 pKa = 10.55ALIVYY188 pKa = 10.32SKK190 pKa = 11.29DD191 pKa = 3.38DD192 pKa = 3.49VLEE195 pKa = 4.46ADD197 pKa = 4.13EE198 pKa = 4.49MVIHH202 pKa = 7.17IDD204 pKa = 3.74IEE206 pKa = 4.31HH207 pKa = 5.69QRR209 pKa = 11.84IPSLQRR215 pKa = 11.84SRR217 pKa = 11.84CDD219 pKa = 3.05STRR222 pKa = 11.84MHH224 pKa = 7.06DD225 pKa = 3.35VRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 3.31
MM1 pKa = 7.26AQNGTGGGSRR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84NNNNNNSTARR28 pKa = 11.84DD29 pKa = 3.58KK30 pKa = 11.54ALLALTQQVNRR41 pKa = 11.84LANIASSSAPSLQHH55 pKa = 5.37PTFIASKK62 pKa = 9.33KK63 pKa = 9.13CRR65 pKa = 11.84AGYY68 pKa = 8.61TYY70 pKa = 10.49TSLDD74 pKa = 3.31VRR76 pKa = 11.84PTRR79 pKa = 11.84TEE81 pKa = 3.58KK82 pKa = 10.9DD83 pKa = 2.95KK84 pKa = 11.8SFGQRR89 pKa = 11.84LIIPVPVSEE98 pKa = 4.23YY99 pKa = 9.43PKK101 pKa = 10.66KK102 pKa = 10.4KK103 pKa = 9.72VSCVQVRR110 pKa = 11.84LNPSPKK116 pKa = 9.47FNSTIWVSLRR126 pKa = 11.84RR127 pKa = 11.84LDD129 pKa = 3.62EE130 pKa = 4.33TTLLTSEE137 pKa = 4.43NVFKK141 pKa = 11.33LFTDD145 pKa = 4.24GLAAVLIYY153 pKa = 9.4QHH155 pKa = 6.54VPTGIQPNNKK165 pKa = 8.16ITFDD169 pKa = 3.57MSNVGAEE176 pKa = 3.78IGDD179 pKa = 3.54MGKK182 pKa = 10.16YY183 pKa = 10.55ALIVYY188 pKa = 10.32SKK190 pKa = 11.29DD191 pKa = 3.38DD192 pKa = 3.49VLEE195 pKa = 4.46ADD197 pKa = 4.13EE198 pKa = 4.49MVIHH202 pKa = 7.17IDD204 pKa = 3.74IEE206 pKa = 4.31HH207 pKa = 5.69QRR209 pKa = 11.84IPSLQRR215 pKa = 11.84SRR217 pKa = 11.84CDD219 pKa = 3.05STRR222 pKa = 11.84MHH224 pKa = 7.06DD225 pKa = 3.35VRR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 3.31
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2920 |
95 |
993 |
417.1 |
46.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.438 ± 0.771 | 2.397 ± 0.2 |
6.61 ± 0.685 | 5.205 ± 0.34 |
4.041 ± 0.564 | 5.137 ± 0.277 |
2.637 ± 0.247 | 5.034 ± 0.261 |
5.685 ± 0.542 | 9.178 ± 0.324 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.205 | 4.178 ± 0.597 |
4.486 ± 0.642 | 3.699 ± 0.315 |
6.233 ± 0.74 | 9.075 ± 0.807 |
6.712 ± 0.505 | 6.849 ± 0.488 |
0.856 ± 0.162 | 3.082 ± 0.266 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
