
Nannocystis exedens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Nannocystineae; Nannocystaceae; Nannocystis
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
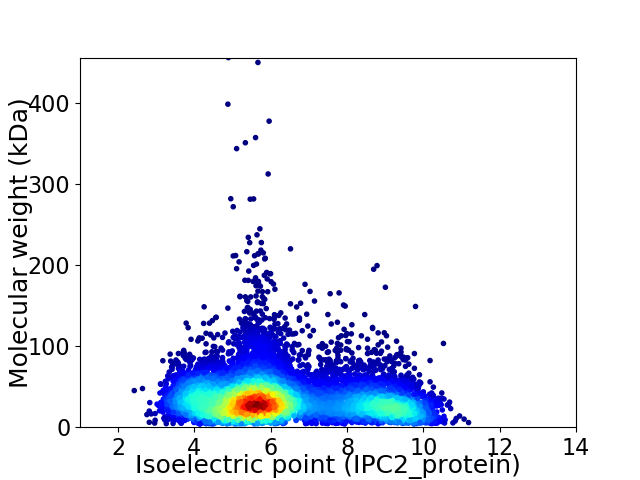
Virtual 2D-PAGE plot for 9143 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
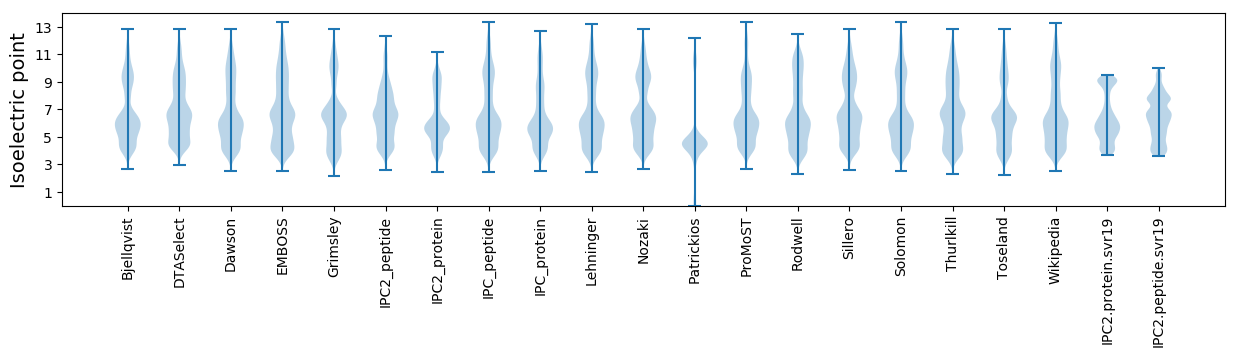
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2FED5|A0A1I2FED5_9DELT NAD-dependent protein deacetylase SIR2 family OS=Nannocystis exedens OX=54 GN=SAMN02745121_06555 PE=4 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84SRR4 pKa = 11.84ISNHH8 pKa = 5.46GPGFRR13 pKa = 11.84SFTTTSTIVAAFAAFAPEE31 pKa = 3.8AHH33 pKa = 7.21ADD35 pKa = 3.68PTRR38 pKa = 11.84LWGTYY43 pKa = 10.09DD44 pKa = 3.48GGSGTDD50 pKa = 2.87AGRR53 pKa = 11.84GVVIDD58 pKa = 4.56DD59 pKa = 4.17EE60 pKa = 5.23FNVVCVGEE68 pKa = 4.56TTSEE72 pKa = 3.86TGITADD78 pKa = 3.6AAHH81 pKa = 7.08DD82 pKa = 3.65GDD84 pKa = 4.6YY85 pKa = 11.45NGGRR89 pKa = 11.84DD90 pKa = 3.55AFITKK95 pKa = 9.93FDD97 pKa = 3.56WFGQQIWGNYY107 pKa = 7.98YY108 pKa = 10.5GGSGDD113 pKa = 5.79DD114 pKa = 3.32IFHH117 pKa = 7.26DD118 pKa = 4.13VVFDD122 pKa = 3.69EE123 pKa = 5.45AEE125 pKa = 4.09GTFFAGGATRR135 pKa = 11.84SANTGNVIATTGTHH149 pKa = 4.59QTALDD154 pKa = 3.87GTGDD158 pKa = 3.33AMLVKK163 pKa = 10.33FRR165 pKa = 11.84NNGARR170 pKa = 11.84LWGTYY175 pKa = 10.08FGGDD179 pKa = 3.31QLEE182 pKa = 4.37EE183 pKa = 4.03AFATCIDD190 pKa = 3.89SQGDD194 pKa = 3.86VYY196 pKa = 11.03IVGTTTSEE204 pKa = 4.0GLFDD208 pKa = 3.88GMVGFTPWDD217 pKa = 3.86DD218 pKa = 4.07EE219 pKa = 4.58LDD221 pKa = 3.71GSIDD225 pKa = 3.42AFIAKK230 pKa = 9.91FDD232 pKa = 3.97GTGGTLLWGTYY243 pKa = 9.56YY244 pKa = 10.73GGSGGEE250 pKa = 4.3TKK252 pKa = 10.76ALDD255 pKa = 3.86CAVGSDD261 pKa = 3.39DD262 pKa = 3.63AVYY265 pKa = 9.15VTGHH269 pKa = 4.98TTSANNEE276 pKa = 3.98LDD278 pKa = 3.32GSHH281 pKa = 7.17FDD283 pKa = 4.04TYY285 pKa = 11.69GGMQDD290 pKa = 3.54AFLVRR295 pKa = 11.84YY296 pKa = 9.38DD297 pKa = 3.54SSGVLEE303 pKa = 3.97WATYY307 pKa = 10.44YY308 pKa = 11.36GDD310 pKa = 3.64TGTDD314 pKa = 3.1QARR317 pKa = 11.84GVAVDD322 pKa = 3.59VSGTDD327 pKa = 3.29DD328 pKa = 3.36LVYY331 pKa = 10.48IVGLTDD337 pKa = 3.39SDD339 pKa = 3.32NDD341 pKa = 3.35IAVNSTTLRR350 pKa = 11.84AGSTDD355 pKa = 3.22AFITQFDD362 pKa = 3.74VDD364 pKa = 5.48GDD366 pKa = 4.32ILWGRR371 pKa = 11.84YY372 pKa = 8.4YY373 pKa = 11.03GGAGLDD379 pKa = 3.37QFEE382 pKa = 4.46GVSAEE387 pKa = 4.11GVIQASGATSSSGLFTSGFDD407 pKa = 3.3NAYY410 pKa = 10.83SGFGWDD416 pKa = 3.52GMFVTFDD423 pKa = 3.68ANGTAVWGSYY433 pKa = 10.39NGGDD437 pKa = 3.64GLDD440 pKa = 3.5EE441 pKa = 4.19SVIAVDD447 pKa = 3.84RR448 pKa = 11.84RR449 pKa = 11.84YY450 pKa = 9.43DD451 pKa = 3.46TLVGVGSTTSTSNTGINDD469 pKa = 3.69SNSPGTIHH477 pKa = 7.8DD478 pKa = 3.49PTYY481 pKa = 11.08NGDD484 pKa = 3.37YY485 pKa = 10.46DD486 pKa = 3.96IFMTLIRR493 pKa = 11.84LFSTT497 pKa = 4.16
MM1 pKa = 8.0RR2 pKa = 11.84SRR4 pKa = 11.84ISNHH8 pKa = 5.46GPGFRR13 pKa = 11.84SFTTTSTIVAAFAAFAPEE31 pKa = 3.8AHH33 pKa = 7.21ADD35 pKa = 3.68PTRR38 pKa = 11.84LWGTYY43 pKa = 10.09DD44 pKa = 3.48GGSGTDD50 pKa = 2.87AGRR53 pKa = 11.84GVVIDD58 pKa = 4.56DD59 pKa = 4.17EE60 pKa = 5.23FNVVCVGEE68 pKa = 4.56TTSEE72 pKa = 3.86TGITADD78 pKa = 3.6AAHH81 pKa = 7.08DD82 pKa = 3.65GDD84 pKa = 4.6YY85 pKa = 11.45NGGRR89 pKa = 11.84DD90 pKa = 3.55AFITKK95 pKa = 9.93FDD97 pKa = 3.56WFGQQIWGNYY107 pKa = 7.98YY108 pKa = 10.5GGSGDD113 pKa = 5.79DD114 pKa = 3.32IFHH117 pKa = 7.26DD118 pKa = 4.13VVFDD122 pKa = 3.69EE123 pKa = 5.45AEE125 pKa = 4.09GTFFAGGATRR135 pKa = 11.84SANTGNVIATTGTHH149 pKa = 4.59QTALDD154 pKa = 3.87GTGDD158 pKa = 3.33AMLVKK163 pKa = 10.33FRR165 pKa = 11.84NNGARR170 pKa = 11.84LWGTYY175 pKa = 10.08FGGDD179 pKa = 3.31QLEE182 pKa = 4.37EE183 pKa = 4.03AFATCIDD190 pKa = 3.89SQGDD194 pKa = 3.86VYY196 pKa = 11.03IVGTTTSEE204 pKa = 4.0GLFDD208 pKa = 3.88GMVGFTPWDD217 pKa = 3.86DD218 pKa = 4.07EE219 pKa = 4.58LDD221 pKa = 3.71GSIDD225 pKa = 3.42AFIAKK230 pKa = 9.91FDD232 pKa = 3.97GTGGTLLWGTYY243 pKa = 9.56YY244 pKa = 10.73GGSGGEE250 pKa = 4.3TKK252 pKa = 10.76ALDD255 pKa = 3.86CAVGSDD261 pKa = 3.39DD262 pKa = 3.63AVYY265 pKa = 9.15VTGHH269 pKa = 4.98TTSANNEE276 pKa = 3.98LDD278 pKa = 3.32GSHH281 pKa = 7.17FDD283 pKa = 4.04TYY285 pKa = 11.69GGMQDD290 pKa = 3.54AFLVRR295 pKa = 11.84YY296 pKa = 9.38DD297 pKa = 3.54SSGVLEE303 pKa = 3.97WATYY307 pKa = 10.44YY308 pKa = 11.36GDD310 pKa = 3.64TGTDD314 pKa = 3.1QARR317 pKa = 11.84GVAVDD322 pKa = 3.59VSGTDD327 pKa = 3.29DD328 pKa = 3.36LVYY331 pKa = 10.48IVGLTDD337 pKa = 3.39SDD339 pKa = 3.32NDD341 pKa = 3.35IAVNSTTLRR350 pKa = 11.84AGSTDD355 pKa = 3.22AFITQFDD362 pKa = 3.74VDD364 pKa = 5.48GDD366 pKa = 4.32ILWGRR371 pKa = 11.84YY372 pKa = 8.4YY373 pKa = 11.03GGAGLDD379 pKa = 3.37QFEE382 pKa = 4.46GVSAEE387 pKa = 4.11GVIQASGATSSSGLFTSGFDD407 pKa = 3.3NAYY410 pKa = 10.83SGFGWDD416 pKa = 3.52GMFVTFDD423 pKa = 3.68ANGTAVWGSYY433 pKa = 10.39NGGDD437 pKa = 3.64GLDD440 pKa = 3.5EE441 pKa = 4.19SVIAVDD447 pKa = 3.84RR448 pKa = 11.84RR449 pKa = 11.84YY450 pKa = 9.43DD451 pKa = 3.46TLVGVGSTTSTSNTGINDD469 pKa = 3.69SNSPGTIHH477 pKa = 7.8DD478 pKa = 3.49PTYY481 pKa = 11.08NGDD484 pKa = 3.37YY485 pKa = 10.46DD486 pKa = 3.96IFMTLIRR493 pKa = 11.84LFSTT497 pKa = 4.16
Molecular weight: 52.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2GC44|A0A1I2GC44_9DELT Undecaprenyl-diphosphatase OS=Nannocystis exedens OX=54 GN=uppP PE=3 SV=1
MM1 pKa = 7.59ARR3 pKa = 11.84PSATPRR9 pKa = 11.84SRR11 pKa = 11.84ILAHH15 pKa = 6.27PRR17 pKa = 11.84RR18 pKa = 11.84ATWMVSRR25 pKa = 11.84WRR27 pKa = 11.84CFSSIARR34 pKa = 11.84ASGNSSRR41 pKa = 11.84GPTHH45 pKa = 6.79HH46 pKa = 7.01VRR48 pKa = 11.84LATPWSLIASIARR61 pKa = 11.84NSLAQPRR68 pKa = 11.84HH69 pKa = 4.74VRR71 pKa = 11.84LATTAVASASIARR84 pKa = 11.84RR85 pKa = 11.84TRR87 pKa = 11.84AVLSS91 pKa = 3.74
MM1 pKa = 7.59ARR3 pKa = 11.84PSATPRR9 pKa = 11.84SRR11 pKa = 11.84ILAHH15 pKa = 6.27PRR17 pKa = 11.84RR18 pKa = 11.84ATWMVSRR25 pKa = 11.84WRR27 pKa = 11.84CFSSIARR34 pKa = 11.84ASGNSSRR41 pKa = 11.84GPTHH45 pKa = 6.79HH46 pKa = 7.01VRR48 pKa = 11.84LATPWSLIASIARR61 pKa = 11.84NSLAQPRR68 pKa = 11.84HH69 pKa = 4.74VRR71 pKa = 11.84LATTAVASASIARR84 pKa = 11.84RR85 pKa = 11.84TRR87 pKa = 11.84AVLSS91 pKa = 3.74
Molecular weight: 9.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3282201 |
39 |
4182 |
359.0 |
38.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.677 ± 0.045 | 1.383 ± 0.017 |
6.225 ± 0.022 | 6.022 ± 0.023 |
3.099 ± 0.015 | 9.029 ± 0.034 |
2.17 ± 0.013 | 3.351 ± 0.016 |
2.229 ± 0.022 | 10.406 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.68 ± 0.01 | 1.824 ± 0.014 |
6.556 ± 0.03 | 2.934 ± 0.014 |
8.045 ± 0.037 | 4.954 ± 0.018 |
5.399 ± 0.026 | 7.632 ± 0.022 |
1.423 ± 0.01 | 1.961 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
