
Rat stool-associated circular ssDNA virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus ratas1
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
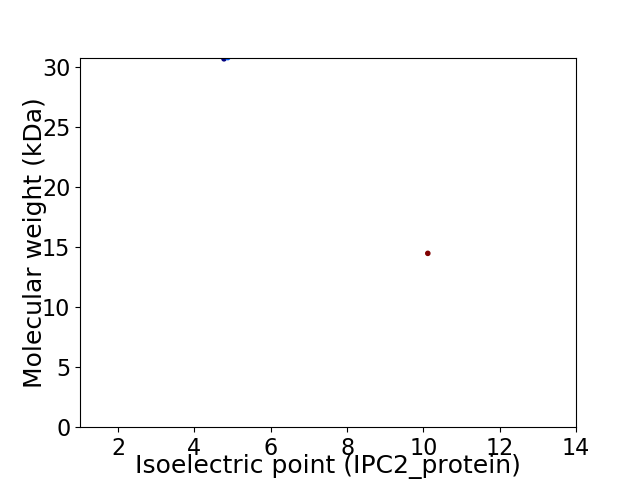
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2LV72|A0A0S2LV72_9VIRU Viral_Rep domain-containing protein OS=Rat stool-associated circular ssDNA virus OX=1699316 PE=4 SV=1
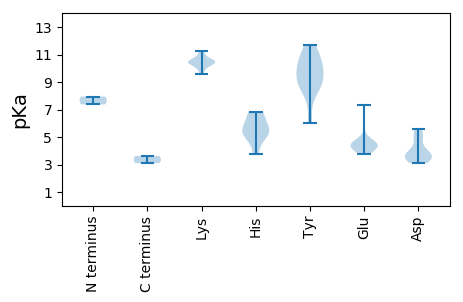
MM1 pKa = 7.37SAADD5 pKa = 3.58MTTAFWGMVINNYY18 pKa = 10.65DD19 pKa = 3.74EE20 pKa = 4.46TDD22 pKa = 3.46LALVQQGYY30 pKa = 8.97PDD32 pKa = 5.33HH33 pKa = 6.71IRR35 pKa = 11.84QMVYY39 pKa = 9.14TLEE42 pKa = 4.8KK43 pKa = 10.5GEE45 pKa = 4.65SGTPHH50 pKa = 5.22IQAYY54 pKa = 8.53VRR56 pKa = 11.84LYY58 pKa = 8.93RR59 pKa = 11.84QQRR62 pKa = 11.84LSYY65 pKa = 10.03VKK67 pKa = 10.43KK68 pKa = 10.22LFPRR72 pKa = 11.84GSFKK76 pKa = 11.11ALLAEE81 pKa = 4.42EE82 pKa = 4.6YY83 pKa = 10.51KK84 pKa = 11.22LNAQRR89 pKa = 11.84YY90 pKa = 6.01AQKK93 pKa = 10.59LDD95 pKa = 3.27KK96 pKa = 9.89TAEE99 pKa = 3.98SSAVITNNPFPDD111 pKa = 4.02PVQEE115 pKa = 3.94LLAVIRR121 pKa = 11.84DD122 pKa = 3.84GFEE125 pKa = 4.1TFHH128 pKa = 6.85EE129 pKa = 4.64SSDD132 pKa = 3.48PAKK135 pKa = 9.55WSKK138 pKa = 10.32YY139 pKa = 8.93DD140 pKa = 3.59KK141 pKa = 10.69RR142 pKa = 11.84DD143 pKa = 3.11VAAWKK148 pKa = 10.53DD149 pKa = 3.45SIEE152 pKa = 4.03RR153 pKa = 11.84VRR155 pKa = 11.84VEE157 pKa = 4.58EE158 pKa = 4.35KK159 pKa = 10.36PNLAKK164 pKa = 10.43FYY166 pKa = 10.92VSPTYY171 pKa = 9.99TRR173 pKa = 11.84VWAKK177 pKa = 10.14FGPSLIRR184 pKa = 11.84HH185 pKa = 5.42VVAQVQSEE193 pKa = 4.3RR194 pKa = 11.84GNSGHH199 pKa = 5.74THH201 pKa = 5.14THH203 pKa = 3.78THH205 pKa = 5.25TDD207 pKa = 3.23EE208 pKa = 4.39KK209 pKa = 10.64FSRR212 pKa = 11.84PGCITINAPSSEE224 pKa = 3.78GSEE227 pKa = 4.32EE228 pKa = 4.08GSEE231 pKa = 4.02ACSPGRR237 pKa = 11.84EE238 pKa = 4.06EE239 pKa = 6.0EE240 pKa = 4.39DD241 pKa = 4.88DD242 pKa = 3.74EE243 pKa = 7.36DD244 pKa = 5.56YY245 pKa = 11.67DD246 pKa = 4.91DD247 pKa = 5.36GSGSEE252 pKa = 4.52TEE254 pKa = 4.67ASTEE258 pKa = 4.25GDD260 pKa = 3.27DD261 pKa = 4.46SGCSEE266 pKa = 4.67SDD268 pKa = 3.9AEE270 pKa = 5.22SEE272 pKa = 4.55DD273 pKa = 3.62
MM1 pKa = 7.37SAADD5 pKa = 3.58MTTAFWGMVINNYY18 pKa = 10.65DD19 pKa = 3.74EE20 pKa = 4.46TDD22 pKa = 3.46LALVQQGYY30 pKa = 8.97PDD32 pKa = 5.33HH33 pKa = 6.71IRR35 pKa = 11.84QMVYY39 pKa = 9.14TLEE42 pKa = 4.8KK43 pKa = 10.5GEE45 pKa = 4.65SGTPHH50 pKa = 5.22IQAYY54 pKa = 8.53VRR56 pKa = 11.84LYY58 pKa = 8.93RR59 pKa = 11.84QQRR62 pKa = 11.84LSYY65 pKa = 10.03VKK67 pKa = 10.43KK68 pKa = 10.22LFPRR72 pKa = 11.84GSFKK76 pKa = 11.11ALLAEE81 pKa = 4.42EE82 pKa = 4.6YY83 pKa = 10.51KK84 pKa = 11.22LNAQRR89 pKa = 11.84YY90 pKa = 6.01AQKK93 pKa = 10.59LDD95 pKa = 3.27KK96 pKa = 9.89TAEE99 pKa = 3.98SSAVITNNPFPDD111 pKa = 4.02PVQEE115 pKa = 3.94LLAVIRR121 pKa = 11.84DD122 pKa = 3.84GFEE125 pKa = 4.1TFHH128 pKa = 6.85EE129 pKa = 4.64SSDD132 pKa = 3.48PAKK135 pKa = 9.55WSKK138 pKa = 10.32YY139 pKa = 8.93DD140 pKa = 3.59KK141 pKa = 10.69RR142 pKa = 11.84DD143 pKa = 3.11VAAWKK148 pKa = 10.53DD149 pKa = 3.45SIEE152 pKa = 4.03RR153 pKa = 11.84VRR155 pKa = 11.84VEE157 pKa = 4.58EE158 pKa = 4.35KK159 pKa = 10.36PNLAKK164 pKa = 10.43FYY166 pKa = 10.92VSPTYY171 pKa = 9.99TRR173 pKa = 11.84VWAKK177 pKa = 10.14FGPSLIRR184 pKa = 11.84HH185 pKa = 5.42VVAQVQSEE193 pKa = 4.3RR194 pKa = 11.84GNSGHH199 pKa = 5.74THH201 pKa = 5.14THH203 pKa = 3.78THH205 pKa = 5.25TDD207 pKa = 3.23EE208 pKa = 4.39KK209 pKa = 10.64FSRR212 pKa = 11.84PGCITINAPSSEE224 pKa = 3.78GSEE227 pKa = 4.32EE228 pKa = 4.08GSEE231 pKa = 4.02ACSPGRR237 pKa = 11.84EE238 pKa = 4.06EE239 pKa = 6.0EE240 pKa = 4.39DD241 pKa = 4.88DD242 pKa = 3.74EE243 pKa = 7.36DD244 pKa = 5.56YY245 pKa = 11.67DD246 pKa = 4.91DD247 pKa = 5.36GSGSEE252 pKa = 4.52TEE254 pKa = 4.67ASTEE258 pKa = 4.25GDD260 pKa = 3.27DD261 pKa = 4.46SGCSEE266 pKa = 4.67SDD268 pKa = 3.9AEE270 pKa = 5.22SEE272 pKa = 4.55DD273 pKa = 3.62
Molecular weight: 30.65 kDa
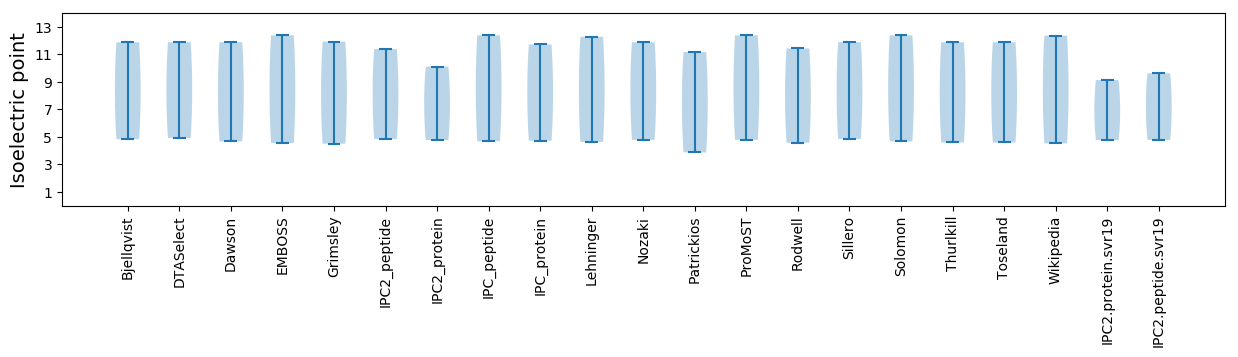
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2LV72|A0A0S2LV72_9VIRU Viral_Rep domain-containing protein OS=Rat stool-associated circular ssDNA virus OX=1699316 PE=4 SV=1
MM1 pKa = 7.95LIMVEE6 pKa = 4.41LRR8 pKa = 11.84PTGIRR13 pKa = 11.84RR14 pKa = 11.84TPCLRR19 pKa = 11.84RR20 pKa = 11.84LFLSLSRR27 pKa = 11.84RR28 pKa = 11.84PSSFTRR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84TFTPRR42 pKa = 11.84ITLRR46 pKa = 11.84CMLSDD51 pKa = 3.77MCLMTRR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84TTSPLTICLVRR72 pKa = 11.84WSMLLRR78 pKa = 11.84ISCGSRR84 pKa = 11.84TISEE88 pKa = 4.74KK89 pKa = 10.45IFSPFIGACFGSHH102 pKa = 6.17QEE104 pKa = 4.0RR105 pKa = 11.84CPVAGNRR112 pKa = 11.84PGGSGSSRR120 pKa = 11.84PSACRR125 pKa = 11.84TQQ127 pKa = 3.1
MM1 pKa = 7.95LIMVEE6 pKa = 4.41LRR8 pKa = 11.84PTGIRR13 pKa = 11.84RR14 pKa = 11.84TPCLRR19 pKa = 11.84RR20 pKa = 11.84LFLSLSRR27 pKa = 11.84RR28 pKa = 11.84PSSFTRR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84TFTPRR42 pKa = 11.84ITLRR46 pKa = 11.84CMLSDD51 pKa = 3.77MCLMTRR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84TTSPLTICLVRR72 pKa = 11.84WSMLLRR78 pKa = 11.84ISCGSRR84 pKa = 11.84TISEE88 pKa = 4.74KK89 pKa = 10.45IFSPFIGACFGSHH102 pKa = 6.17QEE104 pKa = 4.0RR105 pKa = 11.84CPVAGNRR112 pKa = 11.84PGGSGSSRR120 pKa = 11.84PSACRR125 pKa = 11.84TQQ127 pKa = 3.1
Molecular weight: 14.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
400 |
127 |
273 |
200.0 |
22.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 1.92 | 2.75 ± 1.752 |
5.5 ± 2.327 | 7.75 ± 2.66 |
3.75 ± 0.481 | 6.25 ± 0.024 |
2.25 ± 0.722 | 4.25 ± 1.012 |
4.0 ± 1.586 | 7.0 ± 1.987 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 1.098 | 2.25 ± 0.722 |
5.75 ± 1.049 | 3.25 ± 0.827 |
9.5 ± 4.251 | 10.75 ± 0.913 |
7.25 ± 1.086 | 4.75 ± 1.179 |
1.25 ± 0.228 | 3.0 ± 1.481 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
