
Porcine associated porprismacovirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
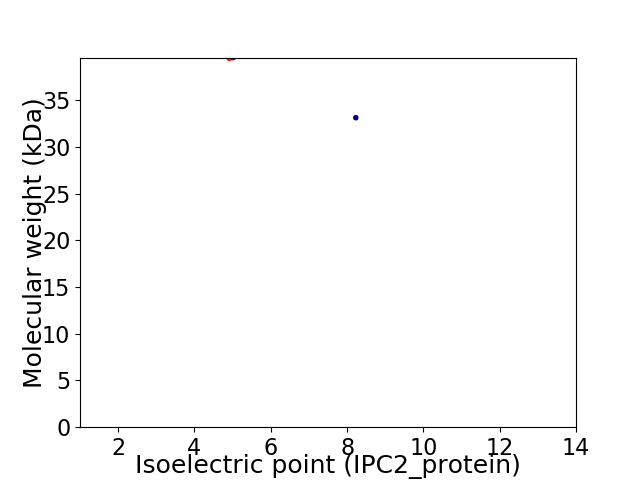
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076VH58|A0A076VH58_9VIRU Rep protein OS=Porcine associated porprismacovirus 6 OX=2170122 PE=4 SV=1
MM1 pKa = 7.13PTGIKK6 pKa = 10.09LALEE10 pKa = 4.28EE11 pKa = 4.47GRR13 pKa = 11.84PTGTGDD19 pKa = 2.94TGGNPMVTVRR29 pKa = 11.84VSEE32 pKa = 4.36TYY34 pKa = 10.72DD35 pKa = 3.26LSTKK39 pKa = 8.75VGKK42 pKa = 9.33MGLVGIHH49 pKa = 6.31TPTGDD54 pKa = 4.83LIPRR58 pKa = 11.84LWSGFVQQYY67 pKa = 9.99KK68 pKa = 10.08KK69 pKa = 10.86FRR71 pKa = 11.84FVKK74 pKa = 10.14CDD76 pKa = 2.95VAMACASMLPADD88 pKa = 4.26PLQIGVEE95 pKa = 4.22SGSIAPQDD103 pKa = 3.58MFNPILYY110 pKa = 9.79KK111 pKa = 10.56AVSNQSMNNFLGWLQLVGEE130 pKa = 4.48TEE132 pKa = 4.22DD133 pKa = 3.65MVGAFNKK140 pKa = 10.41GSVVDD145 pKa = 3.77VNNPVFKK152 pKa = 10.69SAAQDD157 pKa = 3.56GVDD160 pKa = 2.88IDD162 pKa = 3.95QFQMYY167 pKa = 9.94YY168 pKa = 10.9SLLADD173 pKa = 3.27SDD175 pKa = 3.9GWRR178 pKa = 11.84KK179 pKa = 9.97AMPQSGLTMRR189 pKa = 11.84GLIPLVFQVLSNYY202 pKa = 9.75GAGPQNEE209 pKa = 4.57SDD211 pKa = 3.85SADD214 pKa = 3.42LRR216 pKa = 11.84ISVPALSLGPQGGPPTSAADD236 pKa = 3.29SSSFVLPRR244 pKa = 11.84TMRR247 pKa = 11.84GPAARR252 pKa = 11.84MPAIDD257 pKa = 3.43TAFFSSLAAKK267 pKa = 9.89DD268 pKa = 3.73RR269 pKa = 11.84NVFSAFVQAQEE280 pKa = 4.25TLGNVPPAYY289 pKa = 10.26VGIIVLPPAKK299 pKa = 9.97LDD301 pKa = 3.35QLYY304 pKa = 10.56YY305 pKa = 10.03RR306 pKa = 11.84LKK308 pKa = 8.93VTWTIEE314 pKa = 3.99FTGVRR319 pKa = 11.84SNNDD323 pKa = 3.01LSSWSALANAGNVAYY338 pKa = 10.22GSDD341 pKa = 3.73YY342 pKa = 11.36DD343 pKa = 4.06VQSKK347 pKa = 10.82SMASALGMADD357 pKa = 3.23TAGADD362 pKa = 2.99ITKK365 pKa = 10.13IMEE368 pKa = 4.36GSS370 pKa = 3.18
MM1 pKa = 7.13PTGIKK6 pKa = 10.09LALEE10 pKa = 4.28EE11 pKa = 4.47GRR13 pKa = 11.84PTGTGDD19 pKa = 2.94TGGNPMVTVRR29 pKa = 11.84VSEE32 pKa = 4.36TYY34 pKa = 10.72DD35 pKa = 3.26LSTKK39 pKa = 8.75VGKK42 pKa = 9.33MGLVGIHH49 pKa = 6.31TPTGDD54 pKa = 4.83LIPRR58 pKa = 11.84LWSGFVQQYY67 pKa = 9.99KK68 pKa = 10.08KK69 pKa = 10.86FRR71 pKa = 11.84FVKK74 pKa = 10.14CDD76 pKa = 2.95VAMACASMLPADD88 pKa = 4.26PLQIGVEE95 pKa = 4.22SGSIAPQDD103 pKa = 3.58MFNPILYY110 pKa = 9.79KK111 pKa = 10.56AVSNQSMNNFLGWLQLVGEE130 pKa = 4.48TEE132 pKa = 4.22DD133 pKa = 3.65MVGAFNKK140 pKa = 10.41GSVVDD145 pKa = 3.77VNNPVFKK152 pKa = 10.69SAAQDD157 pKa = 3.56GVDD160 pKa = 2.88IDD162 pKa = 3.95QFQMYY167 pKa = 9.94YY168 pKa = 10.9SLLADD173 pKa = 3.27SDD175 pKa = 3.9GWRR178 pKa = 11.84KK179 pKa = 9.97AMPQSGLTMRR189 pKa = 11.84GLIPLVFQVLSNYY202 pKa = 9.75GAGPQNEE209 pKa = 4.57SDD211 pKa = 3.85SADD214 pKa = 3.42LRR216 pKa = 11.84ISVPALSLGPQGGPPTSAADD236 pKa = 3.29SSSFVLPRR244 pKa = 11.84TMRR247 pKa = 11.84GPAARR252 pKa = 11.84MPAIDD257 pKa = 3.43TAFFSSLAAKK267 pKa = 9.89DD268 pKa = 3.73RR269 pKa = 11.84NVFSAFVQAQEE280 pKa = 4.25TLGNVPPAYY289 pKa = 10.26VGIIVLPPAKK299 pKa = 9.97LDD301 pKa = 3.35QLYY304 pKa = 10.56YY305 pKa = 10.03RR306 pKa = 11.84LKK308 pKa = 8.93VTWTIEE314 pKa = 3.99FTGVRR319 pKa = 11.84SNNDD323 pKa = 3.01LSSWSALANAGNVAYY338 pKa = 10.22GSDD341 pKa = 3.73YY342 pKa = 11.36DD343 pKa = 4.06VQSKK347 pKa = 10.82SMASALGMADD357 pKa = 3.23TAGADD362 pKa = 2.99ITKK365 pKa = 10.13IMEE368 pKa = 4.36GSS370 pKa = 3.18
Molecular weight: 39.45 kDa
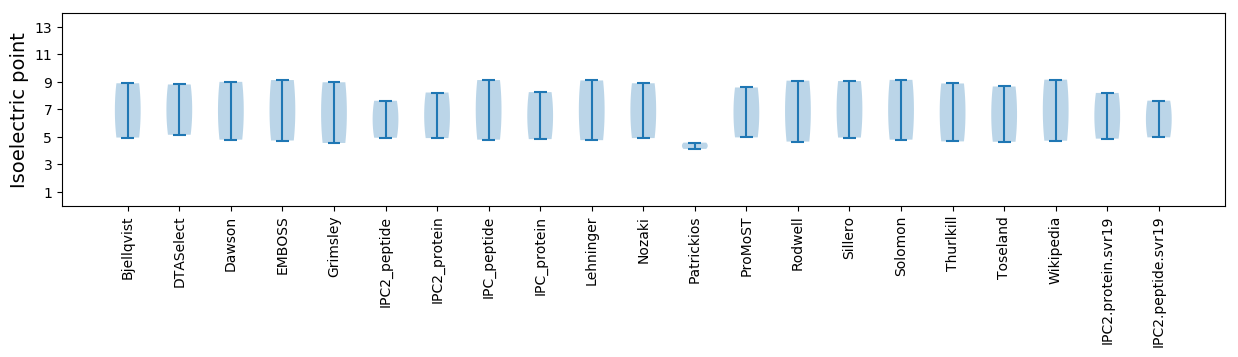
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VH58|A0A076VH58_9VIRU Rep protein OS=Porcine associated porprismacovirus 6 OX=2170122 PE=4 SV=1
MM1 pKa = 6.91KK2 pKa = 9.8TYY4 pKa = 9.96MLTIPRR10 pKa = 11.84EE11 pKa = 3.79RR12 pKa = 11.84ADD14 pKa = 4.56RR15 pKa = 11.84IGTEE19 pKa = 3.92PIIRR23 pKa = 11.84LMRR26 pKa = 11.84YY27 pKa = 8.86IRR29 pKa = 11.84EE30 pKa = 3.98NDD32 pKa = 3.18VKK34 pKa = 10.47KK35 pKa = 9.99WIVAMEE41 pKa = 4.36TGRR44 pKa = 11.84RR45 pKa = 11.84GYY47 pKa = 7.87EE48 pKa = 3.43HH49 pKa = 6.04WQVRR53 pKa = 11.84LQARR57 pKa = 11.84EE58 pKa = 4.23GFSEE62 pKa = 4.17WKK64 pKa = 8.98DD65 pKa = 3.4TEE67 pKa = 4.44VPVLGPNWTIVTEE80 pKa = 4.14KK81 pKa = 10.67QKK83 pKa = 11.34VGVGWIQRR91 pKa = 11.84NIPQAHH97 pKa = 5.73VEE99 pKa = 4.16EE100 pKa = 5.68CSDD103 pKa = 3.01NWEE106 pKa = 4.2YY107 pKa = 10.43EE108 pKa = 4.27AKK110 pKa = 10.26EE111 pKa = 3.95GRR113 pKa = 11.84YY114 pKa = 6.59WASWDD119 pKa = 3.49TVEE122 pKa = 4.17VHH124 pKa = 6.09RR125 pKa = 11.84MRR127 pKa = 11.84WGRR130 pKa = 11.84PRR132 pKa = 11.84WYY134 pKa = 10.17QQALLNKK141 pKa = 9.86LRR143 pKa = 11.84TTTDD147 pKa = 2.96RR148 pKa = 11.84EE149 pKa = 4.18VMVWYY154 pKa = 10.05DD155 pKa = 3.34PEE157 pKa = 4.85GNNGKK162 pKa = 9.23SWLVGHH168 pKa = 7.02LFEE171 pKa = 5.21TGRR174 pKa = 11.84AYY176 pKa = 10.74YY177 pKa = 10.16LPPTLTSVQSMLQTMASLAHH197 pKa = 5.75QDD199 pKa = 3.52RR200 pKa = 11.84EE201 pKa = 4.06KK202 pKa = 10.98GYY204 pKa = 9.13PPKK207 pKa = 10.09PYY209 pKa = 10.45VIIDD213 pKa = 3.76IPRR216 pKa = 11.84TWKK219 pKa = 9.02WSKK222 pKa = 10.19EE223 pKa = 3.98LYY225 pKa = 10.17CAIEE229 pKa = 4.3SIKK232 pKa = 10.82DD233 pKa = 3.66GLIMDD238 pKa = 4.71PRR240 pKa = 11.84YY241 pKa = 9.42SARR244 pKa = 11.84PINIRR249 pKa = 11.84GVKK252 pKa = 10.09VLVLTNEE259 pKa = 4.08MPKK262 pKa = 10.41LDD264 pKa = 4.49ALSEE268 pKa = 4.11DD269 pKa = 2.96RR270 pKa = 11.84WIIEE274 pKa = 4.21NTPMHH279 pKa = 6.64
MM1 pKa = 6.91KK2 pKa = 9.8TYY4 pKa = 9.96MLTIPRR10 pKa = 11.84EE11 pKa = 3.79RR12 pKa = 11.84ADD14 pKa = 4.56RR15 pKa = 11.84IGTEE19 pKa = 3.92PIIRR23 pKa = 11.84LMRR26 pKa = 11.84YY27 pKa = 8.86IRR29 pKa = 11.84EE30 pKa = 3.98NDD32 pKa = 3.18VKK34 pKa = 10.47KK35 pKa = 9.99WIVAMEE41 pKa = 4.36TGRR44 pKa = 11.84RR45 pKa = 11.84GYY47 pKa = 7.87EE48 pKa = 3.43HH49 pKa = 6.04WQVRR53 pKa = 11.84LQARR57 pKa = 11.84EE58 pKa = 4.23GFSEE62 pKa = 4.17WKK64 pKa = 8.98DD65 pKa = 3.4TEE67 pKa = 4.44VPVLGPNWTIVTEE80 pKa = 4.14KK81 pKa = 10.67QKK83 pKa = 11.34VGVGWIQRR91 pKa = 11.84NIPQAHH97 pKa = 5.73VEE99 pKa = 4.16EE100 pKa = 5.68CSDD103 pKa = 3.01NWEE106 pKa = 4.2YY107 pKa = 10.43EE108 pKa = 4.27AKK110 pKa = 10.26EE111 pKa = 3.95GRR113 pKa = 11.84YY114 pKa = 6.59WASWDD119 pKa = 3.49TVEE122 pKa = 4.17VHH124 pKa = 6.09RR125 pKa = 11.84MRR127 pKa = 11.84WGRR130 pKa = 11.84PRR132 pKa = 11.84WYY134 pKa = 10.17QQALLNKK141 pKa = 9.86LRR143 pKa = 11.84TTTDD147 pKa = 2.96RR148 pKa = 11.84EE149 pKa = 4.18VMVWYY154 pKa = 10.05DD155 pKa = 3.34PEE157 pKa = 4.85GNNGKK162 pKa = 9.23SWLVGHH168 pKa = 7.02LFEE171 pKa = 5.21TGRR174 pKa = 11.84AYY176 pKa = 10.74YY177 pKa = 10.16LPPTLTSVQSMLQTMASLAHH197 pKa = 5.75QDD199 pKa = 3.52RR200 pKa = 11.84EE201 pKa = 4.06KK202 pKa = 10.98GYY204 pKa = 9.13PPKK207 pKa = 10.09PYY209 pKa = 10.45VIIDD213 pKa = 3.76IPRR216 pKa = 11.84TWKK219 pKa = 9.02WSKK222 pKa = 10.19EE223 pKa = 3.98LYY225 pKa = 10.17CAIEE229 pKa = 4.3SIKK232 pKa = 10.82DD233 pKa = 3.66GLIMDD238 pKa = 4.71PRR240 pKa = 11.84YY241 pKa = 9.42SARR244 pKa = 11.84PINIRR249 pKa = 11.84GVKK252 pKa = 10.09VLVLTNEE259 pKa = 4.08MPKK262 pKa = 10.41LDD264 pKa = 4.49ALSEE268 pKa = 4.11DD269 pKa = 2.96RR270 pKa = 11.84WIIEE274 pKa = 4.21NTPMHH279 pKa = 6.64
Molecular weight: 33.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
649 |
279 |
370 |
324.5 |
36.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.55 ± 1.934 | 0.616 ± 0.067 |
5.701 ± 0.697 | 5.239 ± 2.25 |
2.619 ± 1.273 | 7.858 ± 1.421 |
1.079 ± 0.717 | 5.239 ± 1.051 |
4.777 ± 0.641 | 7.704 ± 0.598 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.16 ± 0.146 | 4.006 ± 0.282 |
6.317 ± 0.15 | 4.16 ± 0.385 |
5.855 ± 2.078 | 6.934 ± 2.001 |
5.855 ± 0.399 | 7.55 ± 0.495 |
3.082 ± 1.535 | 3.698 ± 0.643 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
