
Nonlabens sp. MB-3u-79
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Nonlabens; unclassified Nonlabens
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
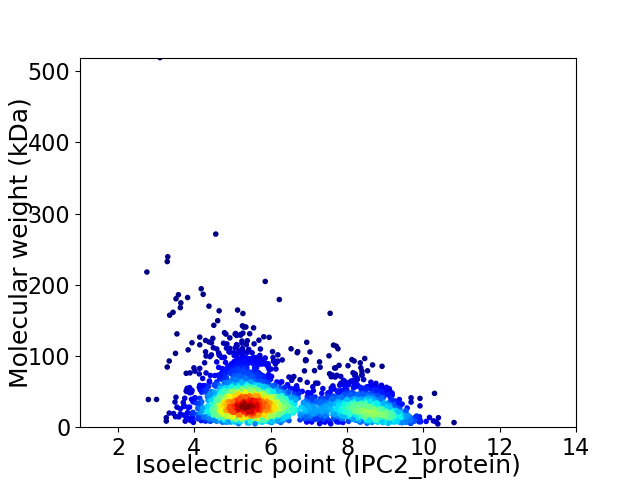
Virtual 2D-PAGE plot for 2710 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
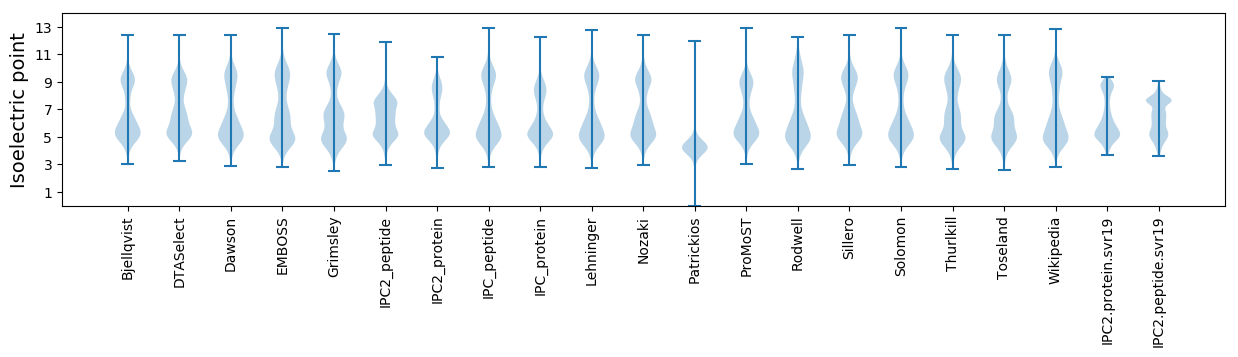
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K8X8N8|A0A2K8X8N8_9FLAO DUF2179 domain-containing protein OS=Nonlabens sp. MB-3u-79 OX=2058134 GN=CW736_07850 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.41KK3 pKa = 10.47LYY5 pKa = 9.71TILAALFLTASAFAQAPEE23 pKa = 4.04KK24 pKa = 10.07MSYY27 pKa = 9.19QAVVRR32 pKa = 11.84DD33 pKa = 3.69SGDD36 pKa = 3.42ALVTSQPVGMQISILQTTATGTAVYY61 pKa = 10.71VEE63 pKa = 4.59TQTPTTNVNGLVSLEE78 pKa = 4.09IGTGSVVSGDD88 pKa = 3.49FTSIDD93 pKa = 3.55WSTDD97 pKa = 2.5SYY99 pKa = 11.09FIKK102 pKa = 10.12TEE104 pKa = 3.84TDD106 pKa = 3.15PAGGTTYY113 pKa = 10.35TITGTSQLMSVPFALYY129 pKa = 10.65AKK131 pKa = 9.6TSGNGAGPIGPQGPAGNDD149 pKa = 3.51GATGPIGADD158 pKa = 3.26GAAGTNGTDD167 pKa = 3.46GAQGIQGAQGDD178 pKa = 4.03IGAAGADD185 pKa = 3.91GAQGIQGADD194 pKa = 3.25GATGTTGQQGPAGNDD209 pKa = 3.37GATGQQGLAGNDD221 pKa = 3.5GATGLTGPVGPTGPAGSLGPQGIQGAAGINGTPGIQGAPGTAGAPGVDD269 pKa = 3.8GAAGTNGTDD278 pKa = 3.46GAQGIQGADD287 pKa = 3.25GATGTTGQQGPAGNDD302 pKa = 3.21GTTGQQGLAGNDD314 pKa = 3.5GATGLTGLVGPAGSLGPQGIQGAAGINGTPGIQGATGASGADD356 pKa = 3.7GAAGAPGTAGAPGVDD371 pKa = 3.8GAAGTNGIDD380 pKa = 3.54GTQGIQGATGADD392 pKa = 4.05GATGPSGTNGTQGIQGATGADD413 pKa = 3.8GAQGIQGATGADD425 pKa = 3.8GAQGIQGEE433 pKa = 4.46TGAAGTNATQPTYY446 pKa = 10.18TVNTLYY452 pKa = 10.26PEE454 pKa = 4.29LGGYY458 pKa = 9.32VIEE461 pKa = 4.66INADD465 pKa = 3.44GTHH468 pKa = 6.22GLVAAMQDD476 pKa = 3.13QGTSNWYY483 pKa = 8.69QANDD487 pKa = 3.93LLSNAANHH495 pKa = 6.57DD496 pKa = 3.81VNGAKK501 pKa = 10.36FKK503 pKa = 10.62DD504 pKa = 3.15WRR506 pKa = 11.84IPTKK510 pKa = 10.63RR511 pKa = 11.84EE512 pKa = 3.36LNLIYY517 pKa = 9.88FQRR520 pKa = 11.84ASIGAASYY528 pKa = 9.25YY529 pKa = 9.58WSSTEE534 pKa = 3.56IDD536 pKa = 4.89NYY538 pKa = 9.06SAWYY542 pKa = 10.23QFFPNGNQGYY552 pKa = 7.77SFKK555 pKa = 11.14SSTEE559 pKa = 3.5YY560 pKa = 10.6VRR562 pKa = 11.84AVRR565 pKa = 11.84AFF567 pKa = 3.27
MM1 pKa = 7.45KK2 pKa = 10.41KK3 pKa = 10.47LYY5 pKa = 9.71TILAALFLTASAFAQAPEE23 pKa = 4.04KK24 pKa = 10.07MSYY27 pKa = 9.19QAVVRR32 pKa = 11.84DD33 pKa = 3.69SGDD36 pKa = 3.42ALVTSQPVGMQISILQTTATGTAVYY61 pKa = 10.71VEE63 pKa = 4.59TQTPTTNVNGLVSLEE78 pKa = 4.09IGTGSVVSGDD88 pKa = 3.49FTSIDD93 pKa = 3.55WSTDD97 pKa = 2.5SYY99 pKa = 11.09FIKK102 pKa = 10.12TEE104 pKa = 3.84TDD106 pKa = 3.15PAGGTTYY113 pKa = 10.35TITGTSQLMSVPFALYY129 pKa = 10.65AKK131 pKa = 9.6TSGNGAGPIGPQGPAGNDD149 pKa = 3.51GATGPIGADD158 pKa = 3.26GAAGTNGTDD167 pKa = 3.46GAQGIQGAQGDD178 pKa = 4.03IGAAGADD185 pKa = 3.91GAQGIQGADD194 pKa = 3.25GATGTTGQQGPAGNDD209 pKa = 3.37GATGQQGLAGNDD221 pKa = 3.5GATGLTGPVGPTGPAGSLGPQGIQGAAGINGTPGIQGAPGTAGAPGVDD269 pKa = 3.8GAAGTNGTDD278 pKa = 3.46GAQGIQGADD287 pKa = 3.25GATGTTGQQGPAGNDD302 pKa = 3.21GTTGQQGLAGNDD314 pKa = 3.5GATGLTGLVGPAGSLGPQGIQGAAGINGTPGIQGATGASGADD356 pKa = 3.7GAAGAPGTAGAPGVDD371 pKa = 3.8GAAGTNGIDD380 pKa = 3.54GTQGIQGATGADD392 pKa = 4.05GATGPSGTNGTQGIQGATGADD413 pKa = 3.8GAQGIQGATGADD425 pKa = 3.8GAQGIQGEE433 pKa = 4.46TGAAGTNATQPTYY446 pKa = 10.18TVNTLYY452 pKa = 10.26PEE454 pKa = 4.29LGGYY458 pKa = 9.32VIEE461 pKa = 4.66INADD465 pKa = 3.44GTHH468 pKa = 6.22GLVAAMQDD476 pKa = 3.13QGTSNWYY483 pKa = 8.69QANDD487 pKa = 3.93LLSNAANHH495 pKa = 6.57DD496 pKa = 3.81VNGAKK501 pKa = 10.36FKK503 pKa = 10.62DD504 pKa = 3.15WRR506 pKa = 11.84IPTKK510 pKa = 10.63RR511 pKa = 11.84EE512 pKa = 3.36LNLIYY517 pKa = 9.88FQRR520 pKa = 11.84ASIGAASYY528 pKa = 9.25YY529 pKa = 9.58WSSTEE534 pKa = 3.56IDD536 pKa = 4.89NYY538 pKa = 9.06SAWYY542 pKa = 10.23QFFPNGNQGYY552 pKa = 7.77SFKK555 pKa = 11.14SSTEE559 pKa = 3.5YY560 pKa = 10.6VRR562 pKa = 11.84AVRR565 pKa = 11.84AFF567 pKa = 3.27
Molecular weight: 54.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K8X8G8|A0A2K8X8G8_9FLAO Por_Secre_tail domain-containing protein OS=Nonlabens sp. MB-3u-79 OX=2058134 GN=CW736_07495 PE=4 SV=1
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 10.49RR39 pKa = 11.84SGGRR43 pKa = 11.84NDD45 pKa = 3.41SGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.61KK54 pKa = 10.94GGGHH58 pKa = 5.71KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.8FKK68 pKa = 10.97RR69 pKa = 11.84NHH71 pKa = 6.8HH72 pKa = 5.87GVPATVASIEE82 pKa = 4.02YY83 pKa = 10.1DD84 pKa = 3.45PNRR87 pKa = 11.84TAFIALVNYY96 pKa = 9.88QDD98 pKa = 3.41GEE100 pKa = 3.91KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 9.73IVAPNGLKK111 pKa = 10.19VGQNIVSGEE120 pKa = 4.1TATPEE125 pKa = 3.8VGNAMKK131 pKa = 10.7LSQIPLGSIVSCIEE145 pKa = 3.64LRR147 pKa = 11.84PGQGATIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.33YY171 pKa = 9.08ATVKK175 pKa = 9.73MPSGEE180 pKa = 4.05TRR182 pKa = 11.84LILQEE187 pKa = 4.06CLATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.3GYY255 pKa = 7.46RR256 pKa = 11.84TRR258 pKa = 11.84DD259 pKa = 3.23KK260 pKa = 11.38NKK262 pKa = 10.31ASTQYY267 pKa = 10.45IIEE270 pKa = 4.13RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.07KK274 pKa = 9.82
MM1 pKa = 7.54SVRR4 pKa = 11.84KK5 pKa = 9.63LKK7 pKa = 10.48PVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNGYY21 pKa = 9.53DD22 pKa = 4.32AITTDD27 pKa = 3.72KK28 pKa = 11.01PEE30 pKa = 4.36KK31 pKa = 10.55SLLAPIKK38 pKa = 10.49RR39 pKa = 11.84SGGRR43 pKa = 11.84NDD45 pKa = 3.41SGRR48 pKa = 11.84MTMRR52 pKa = 11.84YY53 pKa = 9.61KK54 pKa = 10.94GGGHH58 pKa = 5.71KK59 pKa = 9.74RR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.75RR63 pKa = 11.84IIDD66 pKa = 3.8FKK68 pKa = 10.97RR69 pKa = 11.84NHH71 pKa = 6.8HH72 pKa = 5.87GVPATVASIEE82 pKa = 4.02YY83 pKa = 10.1DD84 pKa = 3.45PNRR87 pKa = 11.84TAFIALVNYY96 pKa = 9.88QDD98 pKa = 3.41GEE100 pKa = 3.91KK101 pKa = 10.21RR102 pKa = 11.84YY103 pKa = 9.73IVAPNGLKK111 pKa = 10.19VGQNIVSGEE120 pKa = 4.1TATPEE125 pKa = 3.8VGNAMKK131 pKa = 10.7LSQIPLGSIVSCIEE145 pKa = 3.64LRR147 pKa = 11.84PGQGATIARR156 pKa = 11.84SAGSFAQLMARR167 pKa = 11.84DD168 pKa = 3.96GKK170 pKa = 10.33YY171 pKa = 9.08ATVKK175 pKa = 9.73MPSGEE180 pKa = 4.05TRR182 pKa = 11.84LILQEE187 pKa = 4.06CLATIGAVSNSDD199 pKa = 3.15HH200 pKa = 6.2QLVVSGKK207 pKa = 9.63AGRR210 pKa = 11.84SRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84PVVMNPVDD229 pKa = 3.47HH230 pKa = 7.05PMGGGEE236 pKa = 4.28GKK238 pKa = 10.51SSGGHH243 pKa = 4.59PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.3GYY255 pKa = 7.46RR256 pKa = 11.84TRR258 pKa = 11.84DD259 pKa = 3.23KK260 pKa = 11.38NKK262 pKa = 10.31ASTQYY267 pKa = 10.45IIEE270 pKa = 4.13RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.07KK274 pKa = 9.82
Molecular weight: 29.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
909226 |
38 |
4979 |
335.5 |
37.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.916 ± 0.049 | 0.771 ± 0.026 |
5.925 ± 0.044 | 6.43 ± 0.055 |
4.992 ± 0.043 | 6.525 ± 0.05 |
1.861 ± 0.024 | 7.602 ± 0.045 |
6.924 ± 0.072 | 9.324 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.377 ± 0.026 | 5.617 ± 0.05 |
3.424 ± 0.028 | 3.709 ± 0.028 |
3.698 ± 0.034 | 6.578 ± 0.037 |
5.94 ± 0.058 | 6.325 ± 0.037 |
1.07 ± 0.016 | 3.991 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
