
Turnip crinkle virus (TCV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Betacarmovirus
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
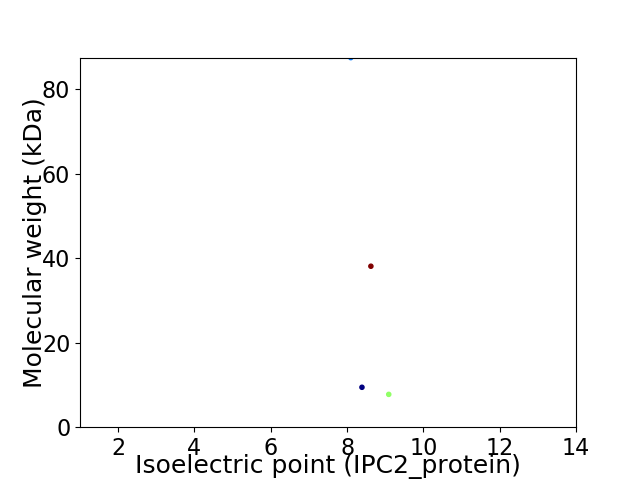
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P17461|MP1_TCV Double gene block protein 1 OS=Turnip crinkle virus OX=11988 GN=ORF2 PE=1 SV=1
MM1 pKa = 7.75PLLHH5 pKa = 6.57TLNTALAVGLLGARR19 pKa = 11.84YY20 pKa = 8.67YY21 pKa = 10.55PEE23 pKa = 3.87VQTFLGLPDD32 pKa = 3.87YY33 pKa = 10.32VGHH36 pKa = 6.28MKK38 pKa = 10.61NVVRR42 pKa = 11.84SVFQGSGLVVVSSDD56 pKa = 3.32TVGVRR61 pKa = 11.84GTYY64 pKa = 9.02SNRR67 pKa = 11.84GQIGSSLGCILAVPDD82 pKa = 3.83SGADD86 pKa = 3.32IEE88 pKa = 4.6IDD90 pKa = 3.42LDD92 pKa = 3.78RR93 pKa = 11.84LVGTEE98 pKa = 4.57EE99 pKa = 4.08EE100 pKa = 4.31ATSCLVEE107 pKa = 4.35AVGSTADD114 pKa = 3.29VPRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84VRR121 pKa = 11.84QKK123 pKa = 10.95GRR125 pKa = 11.84FAMHH129 pKa = 6.97AVNAAKK135 pKa = 10.38LHH137 pKa = 5.72FCGVPKK143 pKa = 9.16PTEE146 pKa = 3.93ANRR149 pKa = 11.84LAVSKK154 pKa = 10.24WLVQYY159 pKa = 10.57CKK161 pKa = 9.79EE162 pKa = 3.85RR163 pKa = 11.84HH164 pKa = 5.4VVDD167 pKa = 2.94SHH169 pKa = 7.07IRR171 pKa = 11.84TIVNTALPRR180 pKa = 11.84VFTPDD185 pKa = 3.26AEE187 pKa = 5.01DD188 pKa = 3.39IQVVLDD194 pKa = 3.55LHH196 pKa = 6.24SVRR199 pKa = 11.84AHH201 pKa = 5.5DD202 pKa = 4.37HH203 pKa = 6.13RR204 pKa = 11.84NALAEE209 pKa = 4.24AGKK212 pKa = 9.78VRR214 pKa = 11.84KK215 pKa = 8.39WWVNLAMHH223 pKa = 7.07PMTGRR228 pKa = 11.84SWSRR232 pKa = 11.84AWRR235 pKa = 11.84RR236 pKa = 11.84LCRR239 pKa = 11.84LPDD242 pKa = 3.68DD243 pKa = 3.79QAISFVRR250 pKa = 11.84XGCLRR255 pKa = 11.84EE256 pKa = 3.88LVGRR260 pKa = 11.84EE261 pKa = 3.84TQISRR266 pKa = 11.84GEE268 pKa = 4.04NPAMRR273 pKa = 11.84VFPLANPPKK282 pKa = 9.64VRR284 pKa = 11.84RR285 pKa = 11.84IFHH288 pKa = 5.85ICGMGNGLDD297 pKa = 3.67FGVHH301 pKa = 5.81NNSLNNLRR309 pKa = 11.84RR310 pKa = 11.84GLMEE314 pKa = 3.52RR315 pKa = 11.84VFYY318 pKa = 11.46VEE320 pKa = 5.45DD321 pKa = 3.56AQKK324 pKa = 10.44QLKK327 pKa = 8.73PAPQPIPGIFGKK339 pKa = 10.79LSGIRR344 pKa = 11.84RR345 pKa = 11.84RR346 pKa = 11.84LVRR349 pKa = 11.84LAGNHH354 pKa = 4.98TPVPRR359 pKa = 11.84EE360 pKa = 4.13KK361 pKa = 10.99YY362 pKa = 8.25PSFYY366 pKa = 10.14KK367 pKa = 10.54GRR369 pKa = 11.84RR370 pKa = 11.84ATIYY374 pKa = 10.56QKK376 pKa = 11.35ALDD379 pKa = 4.16SLHH382 pKa = 6.9DD383 pKa = 3.83RR384 pKa = 11.84PVSRR388 pKa = 11.84KK389 pKa = 9.37DD390 pKa = 3.43AEE392 pKa = 4.12LKK394 pKa = 10.12TFVKK398 pKa = 10.54AEE400 pKa = 4.43KK401 pKa = 10.48INFTAKK407 pKa = 10.05KK408 pKa = 9.71DD409 pKa = 3.4PAPRR413 pKa = 11.84VIQPRR418 pKa = 11.84DD419 pKa = 3.13PRR421 pKa = 11.84YY422 pKa = 9.34NIEE425 pKa = 3.65VGKK428 pKa = 9.55YY429 pKa = 7.77LKK431 pKa = 10.27PYY433 pKa = 8.5EE434 pKa = 4.11HH435 pKa = 6.95HH436 pKa = 7.36LYY438 pKa = 10.4RR439 pKa = 11.84AIDD442 pKa = 3.81AMWGGPTVLKK452 pKa = 10.64GYY454 pKa = 10.56DD455 pKa = 3.27VGEE458 pKa = 4.7LGNIMSNTWDD468 pKa = 3.73KK469 pKa = 10.79FRR471 pKa = 11.84KK472 pKa = 6.87TCAIGFDD479 pKa = 3.63MKK481 pKa = 10.85RR482 pKa = 11.84FDD484 pKa = 3.31QHH486 pKa = 7.89VSVDD490 pKa = 3.41ALRR493 pKa = 11.84WEE495 pKa = 4.15HH496 pKa = 6.05SVYY499 pKa = 10.69NAGFNCPEE507 pKa = 4.32LAQLLTWQLTNKK519 pKa = 10.03GVGRR523 pKa = 11.84ASDD526 pKa = 4.1GFIKK530 pKa = 10.79YY531 pKa = 9.73QVDD534 pKa = 3.98GCRR537 pKa = 11.84MSGDD541 pKa = 3.42VNTALGNCLLACSITKK557 pKa = 10.61YY558 pKa = 10.55LMKK561 pKa = 10.64GIKK564 pKa = 9.73CKK566 pKa = 10.67LINNGDD572 pKa = 3.67DD573 pKa = 3.17CVLFFEE579 pKa = 5.06ADD581 pKa = 3.06EE582 pKa = 4.16VDD584 pKa = 3.48RR585 pKa = 11.84VRR587 pKa = 11.84EE588 pKa = 4.03RR589 pKa = 11.84LHH591 pKa = 6.74HH592 pKa = 6.35WIDD595 pKa = 4.2FGFQCIAEE603 pKa = 4.24EE604 pKa = 3.91PQYY607 pKa = 10.96EE608 pKa = 4.2LEE610 pKa = 4.43KK611 pKa = 11.27VEE613 pKa = 4.57FCQMSPIFDD622 pKa = 3.7GEE624 pKa = 4.31GWVMVRR630 pKa = 11.84NPRR633 pKa = 11.84VSLSKK638 pKa = 10.67DD639 pKa = 3.38SYY641 pKa = 11.38STTQWANEE649 pKa = 3.56KK650 pKa = 10.51DD651 pKa = 3.47AARR654 pKa = 11.84WLAAIGEE661 pKa = 4.56CGLAIAGGVPVLQSYY676 pKa = 7.5YY677 pKa = 11.16SCLKK681 pKa = 10.78RR682 pKa = 11.84NFGPLAGDD690 pKa = 3.76YY691 pKa = 9.59KK692 pKa = 11.19KK693 pKa = 11.07KK694 pKa = 9.14MQDD697 pKa = 2.83VSFDD701 pKa = 3.28SGFYY705 pKa = 10.15RR706 pKa = 11.84LSKK709 pKa = 10.85NGMRR713 pKa = 11.84GSKK716 pKa = 10.13DD717 pKa = 3.23VSQDD721 pKa = 2.78ARR723 pKa = 11.84FSFYY727 pKa = 10.6RR728 pKa = 11.84GFGYY732 pKa = 8.92TPDD735 pKa = 3.5EE736 pKa = 4.16QEE738 pKa = 4.36ALEE741 pKa = 4.36EE742 pKa = 4.46YY743 pKa = 10.85YY744 pKa = 11.21DD745 pKa = 4.28NLQLLCEE752 pKa = 4.43WDD754 pKa = 3.19PTGYY758 pKa = 10.36KK759 pKa = 10.26EE760 pKa = 4.1EE761 pKa = 4.93LSDD764 pKa = 3.76RR765 pKa = 11.84WILNEE770 pKa = 4.91FPTTLL775 pKa = 3.63
MM1 pKa = 7.75PLLHH5 pKa = 6.57TLNTALAVGLLGARR19 pKa = 11.84YY20 pKa = 8.67YY21 pKa = 10.55PEE23 pKa = 3.87VQTFLGLPDD32 pKa = 3.87YY33 pKa = 10.32VGHH36 pKa = 6.28MKK38 pKa = 10.61NVVRR42 pKa = 11.84SVFQGSGLVVVSSDD56 pKa = 3.32TVGVRR61 pKa = 11.84GTYY64 pKa = 9.02SNRR67 pKa = 11.84GQIGSSLGCILAVPDD82 pKa = 3.83SGADD86 pKa = 3.32IEE88 pKa = 4.6IDD90 pKa = 3.42LDD92 pKa = 3.78RR93 pKa = 11.84LVGTEE98 pKa = 4.57EE99 pKa = 4.08EE100 pKa = 4.31ATSCLVEE107 pKa = 4.35AVGSTADD114 pKa = 3.29VPRR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84VRR121 pKa = 11.84QKK123 pKa = 10.95GRR125 pKa = 11.84FAMHH129 pKa = 6.97AVNAAKK135 pKa = 10.38LHH137 pKa = 5.72FCGVPKK143 pKa = 9.16PTEE146 pKa = 3.93ANRR149 pKa = 11.84LAVSKK154 pKa = 10.24WLVQYY159 pKa = 10.57CKK161 pKa = 9.79EE162 pKa = 3.85RR163 pKa = 11.84HH164 pKa = 5.4VVDD167 pKa = 2.94SHH169 pKa = 7.07IRR171 pKa = 11.84TIVNTALPRR180 pKa = 11.84VFTPDD185 pKa = 3.26AEE187 pKa = 5.01DD188 pKa = 3.39IQVVLDD194 pKa = 3.55LHH196 pKa = 6.24SVRR199 pKa = 11.84AHH201 pKa = 5.5DD202 pKa = 4.37HH203 pKa = 6.13RR204 pKa = 11.84NALAEE209 pKa = 4.24AGKK212 pKa = 9.78VRR214 pKa = 11.84KK215 pKa = 8.39WWVNLAMHH223 pKa = 7.07PMTGRR228 pKa = 11.84SWSRR232 pKa = 11.84AWRR235 pKa = 11.84RR236 pKa = 11.84LCRR239 pKa = 11.84LPDD242 pKa = 3.68DD243 pKa = 3.79QAISFVRR250 pKa = 11.84XGCLRR255 pKa = 11.84EE256 pKa = 3.88LVGRR260 pKa = 11.84EE261 pKa = 3.84TQISRR266 pKa = 11.84GEE268 pKa = 4.04NPAMRR273 pKa = 11.84VFPLANPPKK282 pKa = 9.64VRR284 pKa = 11.84RR285 pKa = 11.84IFHH288 pKa = 5.85ICGMGNGLDD297 pKa = 3.67FGVHH301 pKa = 5.81NNSLNNLRR309 pKa = 11.84RR310 pKa = 11.84GLMEE314 pKa = 3.52RR315 pKa = 11.84VFYY318 pKa = 11.46VEE320 pKa = 5.45DD321 pKa = 3.56AQKK324 pKa = 10.44QLKK327 pKa = 8.73PAPQPIPGIFGKK339 pKa = 10.79LSGIRR344 pKa = 11.84RR345 pKa = 11.84RR346 pKa = 11.84LVRR349 pKa = 11.84LAGNHH354 pKa = 4.98TPVPRR359 pKa = 11.84EE360 pKa = 4.13KK361 pKa = 10.99YY362 pKa = 8.25PSFYY366 pKa = 10.14KK367 pKa = 10.54GRR369 pKa = 11.84RR370 pKa = 11.84ATIYY374 pKa = 10.56QKK376 pKa = 11.35ALDD379 pKa = 4.16SLHH382 pKa = 6.9DD383 pKa = 3.83RR384 pKa = 11.84PVSRR388 pKa = 11.84KK389 pKa = 9.37DD390 pKa = 3.43AEE392 pKa = 4.12LKK394 pKa = 10.12TFVKK398 pKa = 10.54AEE400 pKa = 4.43KK401 pKa = 10.48INFTAKK407 pKa = 10.05KK408 pKa = 9.71DD409 pKa = 3.4PAPRR413 pKa = 11.84VIQPRR418 pKa = 11.84DD419 pKa = 3.13PRR421 pKa = 11.84YY422 pKa = 9.34NIEE425 pKa = 3.65VGKK428 pKa = 9.55YY429 pKa = 7.77LKK431 pKa = 10.27PYY433 pKa = 8.5EE434 pKa = 4.11HH435 pKa = 6.95HH436 pKa = 7.36LYY438 pKa = 10.4RR439 pKa = 11.84AIDD442 pKa = 3.81AMWGGPTVLKK452 pKa = 10.64GYY454 pKa = 10.56DD455 pKa = 3.27VGEE458 pKa = 4.7LGNIMSNTWDD468 pKa = 3.73KK469 pKa = 10.79FRR471 pKa = 11.84KK472 pKa = 6.87TCAIGFDD479 pKa = 3.63MKK481 pKa = 10.85RR482 pKa = 11.84FDD484 pKa = 3.31QHH486 pKa = 7.89VSVDD490 pKa = 3.41ALRR493 pKa = 11.84WEE495 pKa = 4.15HH496 pKa = 6.05SVYY499 pKa = 10.69NAGFNCPEE507 pKa = 4.32LAQLLTWQLTNKK519 pKa = 10.03GVGRR523 pKa = 11.84ASDD526 pKa = 4.1GFIKK530 pKa = 10.79YY531 pKa = 9.73QVDD534 pKa = 3.98GCRR537 pKa = 11.84MSGDD541 pKa = 3.42VNTALGNCLLACSITKK557 pKa = 10.61YY558 pKa = 10.55LMKK561 pKa = 10.64GIKK564 pKa = 9.73CKK566 pKa = 10.67LINNGDD572 pKa = 3.67DD573 pKa = 3.17CVLFFEE579 pKa = 5.06ADD581 pKa = 3.06EE582 pKa = 4.16VDD584 pKa = 3.48RR585 pKa = 11.84VRR587 pKa = 11.84EE588 pKa = 4.03RR589 pKa = 11.84LHH591 pKa = 6.74HH592 pKa = 6.35WIDD595 pKa = 4.2FGFQCIAEE603 pKa = 4.24EE604 pKa = 3.91PQYY607 pKa = 10.96EE608 pKa = 4.2LEE610 pKa = 4.43KK611 pKa = 11.27VEE613 pKa = 4.57FCQMSPIFDD622 pKa = 3.7GEE624 pKa = 4.31GWVMVRR630 pKa = 11.84NPRR633 pKa = 11.84VSLSKK638 pKa = 10.67DD639 pKa = 3.38SYY641 pKa = 11.38STTQWANEE649 pKa = 3.56KK650 pKa = 10.51DD651 pKa = 3.47AARR654 pKa = 11.84WLAAIGEE661 pKa = 4.56CGLAIAGGVPVLQSYY676 pKa = 7.5YY677 pKa = 11.16SCLKK681 pKa = 10.78RR682 pKa = 11.84NFGPLAGDD690 pKa = 3.76YY691 pKa = 9.59KK692 pKa = 11.19KK693 pKa = 11.07KK694 pKa = 9.14MQDD697 pKa = 2.83VSFDD701 pKa = 3.28SGFYY705 pKa = 10.15RR706 pKa = 11.84LSKK709 pKa = 10.85NGMRR713 pKa = 11.84GSKK716 pKa = 10.13DD717 pKa = 3.23VSQDD721 pKa = 2.78ARR723 pKa = 11.84FSFYY727 pKa = 10.6RR728 pKa = 11.84GFGYY732 pKa = 8.92TPDD735 pKa = 3.5EE736 pKa = 4.16QEE738 pKa = 4.36ALEE741 pKa = 4.36EE742 pKa = 4.46YY743 pKa = 10.85YY744 pKa = 11.21DD745 pKa = 4.28NLQLLCEE752 pKa = 4.43WDD754 pKa = 3.19PTGYY758 pKa = 10.36KK759 pKa = 10.26EE760 pKa = 4.1EE761 pKa = 4.93LSDD764 pKa = 3.76RR765 pKa = 11.84WILNEE770 pKa = 4.91FPTTLL775 pKa = 3.63
Molecular weight: 87.54 kDa
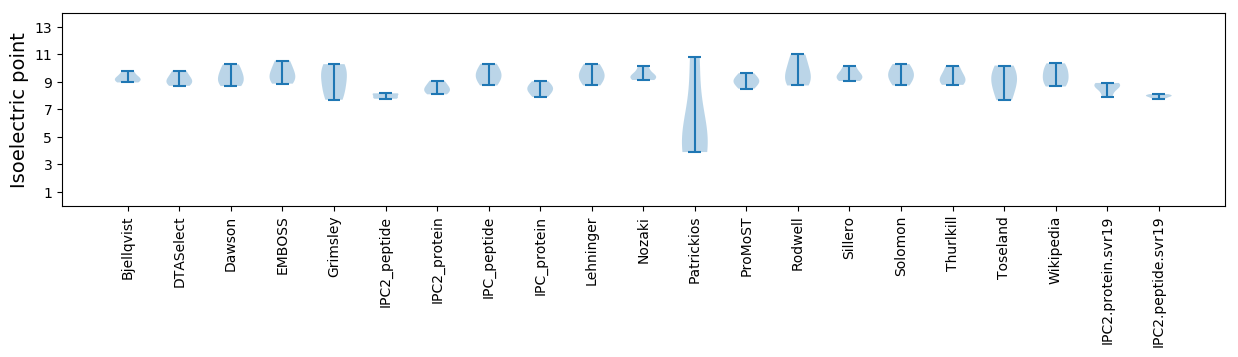
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q7TD19|MP2_TCV Double gene block protein 2 OS=Turnip crinkle virus OX=11988 GN=ORF3 PE=1 SV=2
MM1 pKa = 7.84DD2 pKa = 5.49PEE4 pKa = 5.01RR5 pKa = 11.84IPYY8 pKa = 10.21NSLSDD13 pKa = 3.4SDD15 pKa = 3.7ATGKK19 pKa = 10.03RR20 pKa = 11.84KK21 pKa = 10.17KK22 pKa = 10.59GGEE25 pKa = 3.67KK26 pKa = 9.78SAKK29 pKa = 8.46KK30 pKa = 10.27RR31 pKa = 11.84LVASHH36 pKa = 6.8AASSVLNKK44 pKa = 9.74KK45 pKa = 10.15RR46 pKa = 11.84NEE48 pKa = 3.88GSASHH53 pKa = 6.64GGTWVIVADD62 pKa = 3.93KK63 pKa = 11.25VEE65 pKa = 3.93VSINFNFF72 pKa = 3.74
MM1 pKa = 7.84DD2 pKa = 5.49PEE4 pKa = 5.01RR5 pKa = 11.84IPYY8 pKa = 10.21NSLSDD13 pKa = 3.4SDD15 pKa = 3.7ATGKK19 pKa = 10.03RR20 pKa = 11.84KK21 pKa = 10.17KK22 pKa = 10.59GGEE25 pKa = 3.67KK26 pKa = 9.78SAKK29 pKa = 8.46KK30 pKa = 10.27RR31 pKa = 11.84LVASHH36 pKa = 6.8AASSVLNKK44 pKa = 9.74KK45 pKa = 10.15RR46 pKa = 11.84NEE48 pKa = 3.88GSASHH53 pKa = 6.64GGTWVIVADD62 pKa = 3.93KK63 pKa = 11.25VEE65 pKa = 3.93VSINFNFF72 pKa = 3.74
Molecular weight: 7.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1283 |
72 |
775 |
320.8 |
35.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.716 ± 1.156 | 1.871 ± 0.654 |
5.534 ± 0.594 | 4.988 ± 0.501 |
3.741 ± 0.439 | 7.95 ± 0.399 |
2.182 ± 0.644 | 4.053 ± 0.548 |
6.469 ± 1.235 | 8.496 ± 1.114 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.871 ± 0.283 | 4.131 ± 0.682 |
4.91 ± 0.4 | 3.741 ± 0.757 |
6.625 ± 1.257 | 7.093 ± 1.843 |
5.612 ± 1.551 | 7.716 ± 0.294 |
2.027 ± 0.144 | 3.196 ± 0.458 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
