
candidate division MSBL1 archaeon SCGC-AAA382C18
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Euryarchaeota incertae sedis; candidate division MSBL1
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
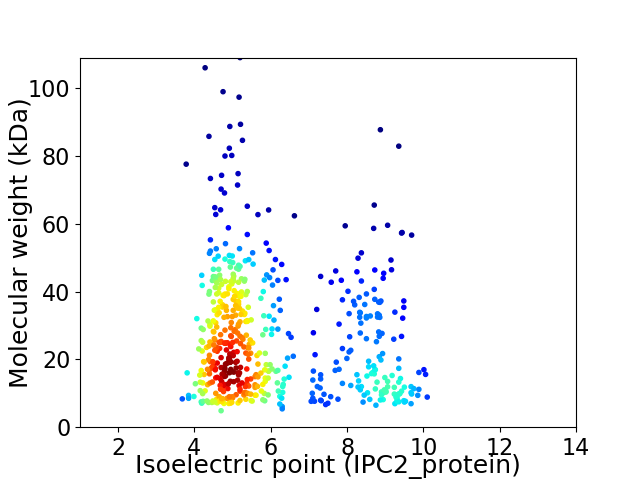
Virtual 2D-PAGE plot for 551 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A133VKL2|A0A133VKL2_9EURY Citramalate synthase (Fragment) OS=candidate division MSBL1 archaeon SCGC-AAA382C18 OX=1698281 GN=AKJ52_01195 PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.18KK3 pKa = 9.61IKK5 pKa = 10.24EE6 pKa = 3.99YY7 pKa = 11.21VIFLLCAMFLSGLVGFFNIDD27 pKa = 2.86AVEE30 pKa = 4.31VVNEE34 pKa = 3.9TAIPPFYY41 pKa = 10.54NRR43 pKa = 11.84PEE45 pKa = 5.07AGDD48 pKa = 3.55SGFLAEE54 pKa = 5.9ANTEE58 pKa = 4.4DD59 pKa = 3.32ITPFDD64 pKa = 3.62EE65 pKa = 4.36MSNVVAPHH73 pKa = 6.59FDD75 pKa = 3.81DD76 pKa = 5.82VITGQSDD83 pKa = 4.02TPTGNSYY90 pKa = 9.71MLFEE94 pKa = 4.21EE95 pKa = 5.52HH96 pKa = 6.78GGDD99 pKa = 3.01WADD102 pKa = 4.92AEE104 pKa = 4.4KK105 pKa = 10.96LPNNDD110 pKa = 4.72EE111 pKa = 5.24DD112 pKa = 6.54DD113 pKa = 4.34LLCWALTASNVLEE126 pKa = 3.86WSGWGFVSDD135 pKa = 5.11FDD137 pKa = 5.4SSDD140 pKa = 3.42DD141 pKa = 3.39MGDD144 pKa = 3.43YY145 pKa = 10.74FEE147 pKa = 5.72SHH149 pKa = 5.33VTDD152 pKa = 3.21SGSWVIYY159 pKa = 9.28GWEE162 pKa = 3.4WWFTGNLPYY171 pKa = 10.94YY172 pKa = 10.52GMGWSYY178 pKa = 11.47EE179 pKa = 4.07DD180 pKa = 4.18EE181 pKa = 4.46NGGDD185 pKa = 3.73FWSDD189 pKa = 2.89EE190 pKa = 4.22YY191 pKa = 11.53DD192 pKa = 3.61YY193 pKa = 11.83SEE195 pKa = 4.29YY196 pKa = 10.61THH198 pKa = 6.63MVDD201 pKa = 4.79NSLKK205 pKa = 9.96TMEE208 pKa = 5.87AIDD211 pKa = 4.26NYY213 pKa = 10.91LHH215 pKa = 6.41NGWGAGLSLGDD226 pKa = 3.95HH227 pKa = 7.57AITVWGFNYY236 pKa = 10.27DD237 pKa = 3.51PSVDD241 pKa = 3.45KK242 pKa = 8.73TTNPHH247 pKa = 7.05DD248 pKa = 4.23YY249 pKa = 11.2YY250 pKa = 11.26LGIWVTDD257 pKa = 3.26SDD259 pKa = 4.77DD260 pKa = 4.55NKK262 pKa = 10.8SQSNPPDD269 pKa = 3.28MLHH272 pKa = 5.48YY273 pKa = 10.94CEE275 pKa = 5.51VEE277 pKa = 3.74WDD279 pKa = 3.61DD280 pKa = 4.95SYY282 pKa = 11.69PGYY285 pKa = 10.93YY286 pKa = 10.44LIPSYY291 pKa = 10.83GRR293 pKa = 11.84SIAMVYY299 pKa = 10.52ALEE302 pKa = 4.46PFPDD306 pKa = 3.67EE307 pKa = 4.2TRR309 pKa = 11.84PTSDD313 pKa = 2.88AGGSYY318 pKa = 9.19TVVEE322 pKa = 4.48GSPVTFDD329 pKa = 4.2ASGSLDD335 pKa = 4.36DD336 pKa = 6.05DD337 pKa = 4.11ALLYY341 pKa = 10.58RR342 pKa = 11.84WDD344 pKa = 4.19LNNDD348 pKa = 3.98RR349 pKa = 11.84VWDD352 pKa = 4.19TSWVSSPTASYY363 pKa = 8.48TWNDD367 pKa = 3.31DD368 pKa = 3.09NYY370 pKa = 11.13GEE372 pKa = 4.36VKK374 pKa = 10.78VEE376 pKa = 4.02VFDD379 pKa = 4.27GRR381 pKa = 11.84LRR383 pKa = 11.84DD384 pKa = 3.31VDD386 pKa = 3.67TATFNIINADD396 pKa = 3.59PTVTATVDD404 pKa = 3.02SDD406 pKa = 4.05TVDD409 pKa = 3.09EE410 pKa = 4.7NGAVTVSGTITDD422 pKa = 3.1SGTRR426 pKa = 11.84DD427 pKa = 3.44NFTVVIDD434 pKa = 3.62WGDD437 pKa = 4.16GISSSYY443 pKa = 10.76SYY445 pKa = 10.36PAGSTDD451 pKa = 3.16YY452 pKa = 11.4SEE454 pKa = 4.0THH456 pKa = 6.31QYY458 pKa = 11.41LDD460 pKa = 4.5DD461 pKa = 4.81NPTEE465 pKa = 4.33TPSDD469 pKa = 3.34NCTINVTVTDD479 pKa = 3.94DD480 pKa = 3.86DD481 pKa = 4.97YY482 pKa = 12.25GVGTANTKK490 pKa = 8.57VTVNNTKK497 pKa = 10.42PMVEE501 pKa = 4.16APFISLQQNSEE512 pKa = 4.3FILPVVHH519 pKa = 5.85EE520 pKa = 4.29TSFVGTFADD529 pKa = 4.09VGTQDD534 pKa = 2.75THH536 pKa = 4.44TAEE539 pKa = 4.37WNWDD543 pKa = 3.58DD544 pKa = 5.28SSTSDD549 pKa = 3.58GTVTEE554 pKa = 4.47SNGLGKK560 pKa = 10.65VSGSHH565 pKa = 5.41TFSSVGEE572 pKa = 3.95YY573 pKa = 10.32TITLTVTDD581 pKa = 5.44DD582 pKa = 3.45DD583 pKa = 4.36TGADD587 pKa = 3.45SSTKK591 pKa = 9.5TICVVDD597 pKa = 3.62AAEE600 pKa = 5.05ALTITNSYY608 pKa = 9.45IQGLSDD614 pKa = 4.74DD615 pKa = 4.29AFKK618 pKa = 11.35SKK620 pKa = 10.95ADD622 pKa = 3.57QRR624 pKa = 11.84KK625 pKa = 8.79KK626 pKa = 10.99AFDD629 pKa = 3.05KK630 pKa = 10.57MFNVLQDD637 pKa = 3.6RR638 pKa = 11.84LADD641 pKa = 3.37EE642 pKa = 5.52HH643 pKa = 6.63YY644 pKa = 10.64RR645 pKa = 11.84AMIQIMRR652 pKa = 11.84NNIRR656 pKa = 11.84AKK658 pKa = 10.76ADD660 pKa = 3.23GSVDD664 pKa = 3.3GKK666 pKa = 11.47VNNDD670 pKa = 3.72WIIDD674 pKa = 3.81TVAQEE679 pKa = 4.43HH680 pKa = 5.84VCQKK684 pKa = 10.11IDD686 pKa = 4.41DD687 pKa = 3.69ITRR690 pKa = 11.84YY691 pKa = 10.19LEE693 pKa = 3.91YY694 pKa = 10.48LIKK697 pKa = 10.73SS698 pKa = 3.68
MM1 pKa = 7.49KK2 pKa = 10.18KK3 pKa = 9.61IKK5 pKa = 10.24EE6 pKa = 3.99YY7 pKa = 11.21VIFLLCAMFLSGLVGFFNIDD27 pKa = 2.86AVEE30 pKa = 4.31VVNEE34 pKa = 3.9TAIPPFYY41 pKa = 10.54NRR43 pKa = 11.84PEE45 pKa = 5.07AGDD48 pKa = 3.55SGFLAEE54 pKa = 5.9ANTEE58 pKa = 4.4DD59 pKa = 3.32ITPFDD64 pKa = 3.62EE65 pKa = 4.36MSNVVAPHH73 pKa = 6.59FDD75 pKa = 3.81DD76 pKa = 5.82VITGQSDD83 pKa = 4.02TPTGNSYY90 pKa = 9.71MLFEE94 pKa = 4.21EE95 pKa = 5.52HH96 pKa = 6.78GGDD99 pKa = 3.01WADD102 pKa = 4.92AEE104 pKa = 4.4KK105 pKa = 10.96LPNNDD110 pKa = 4.72EE111 pKa = 5.24DD112 pKa = 6.54DD113 pKa = 4.34LLCWALTASNVLEE126 pKa = 3.86WSGWGFVSDD135 pKa = 5.11FDD137 pKa = 5.4SSDD140 pKa = 3.42DD141 pKa = 3.39MGDD144 pKa = 3.43YY145 pKa = 10.74FEE147 pKa = 5.72SHH149 pKa = 5.33VTDD152 pKa = 3.21SGSWVIYY159 pKa = 9.28GWEE162 pKa = 3.4WWFTGNLPYY171 pKa = 10.94YY172 pKa = 10.52GMGWSYY178 pKa = 11.47EE179 pKa = 4.07DD180 pKa = 4.18EE181 pKa = 4.46NGGDD185 pKa = 3.73FWSDD189 pKa = 2.89EE190 pKa = 4.22YY191 pKa = 11.53DD192 pKa = 3.61YY193 pKa = 11.83SEE195 pKa = 4.29YY196 pKa = 10.61THH198 pKa = 6.63MVDD201 pKa = 4.79NSLKK205 pKa = 9.96TMEE208 pKa = 5.87AIDD211 pKa = 4.26NYY213 pKa = 10.91LHH215 pKa = 6.41NGWGAGLSLGDD226 pKa = 3.95HH227 pKa = 7.57AITVWGFNYY236 pKa = 10.27DD237 pKa = 3.51PSVDD241 pKa = 3.45KK242 pKa = 8.73TTNPHH247 pKa = 7.05DD248 pKa = 4.23YY249 pKa = 11.2YY250 pKa = 11.26LGIWVTDD257 pKa = 3.26SDD259 pKa = 4.77DD260 pKa = 4.55NKK262 pKa = 10.8SQSNPPDD269 pKa = 3.28MLHH272 pKa = 5.48YY273 pKa = 10.94CEE275 pKa = 5.51VEE277 pKa = 3.74WDD279 pKa = 3.61DD280 pKa = 4.95SYY282 pKa = 11.69PGYY285 pKa = 10.93YY286 pKa = 10.44LIPSYY291 pKa = 10.83GRR293 pKa = 11.84SIAMVYY299 pKa = 10.52ALEE302 pKa = 4.46PFPDD306 pKa = 3.67EE307 pKa = 4.2TRR309 pKa = 11.84PTSDD313 pKa = 2.88AGGSYY318 pKa = 9.19TVVEE322 pKa = 4.48GSPVTFDD329 pKa = 4.2ASGSLDD335 pKa = 4.36DD336 pKa = 6.05DD337 pKa = 4.11ALLYY341 pKa = 10.58RR342 pKa = 11.84WDD344 pKa = 4.19LNNDD348 pKa = 3.98RR349 pKa = 11.84VWDD352 pKa = 4.19TSWVSSPTASYY363 pKa = 8.48TWNDD367 pKa = 3.31DD368 pKa = 3.09NYY370 pKa = 11.13GEE372 pKa = 4.36VKK374 pKa = 10.78VEE376 pKa = 4.02VFDD379 pKa = 4.27GRR381 pKa = 11.84LRR383 pKa = 11.84DD384 pKa = 3.31VDD386 pKa = 3.67TATFNIINADD396 pKa = 3.59PTVTATVDD404 pKa = 3.02SDD406 pKa = 4.05TVDD409 pKa = 3.09EE410 pKa = 4.7NGAVTVSGTITDD422 pKa = 3.1SGTRR426 pKa = 11.84DD427 pKa = 3.44NFTVVIDD434 pKa = 3.62WGDD437 pKa = 4.16GISSSYY443 pKa = 10.76SYY445 pKa = 10.36PAGSTDD451 pKa = 3.16YY452 pKa = 11.4SEE454 pKa = 4.0THH456 pKa = 6.31QYY458 pKa = 11.41LDD460 pKa = 4.5DD461 pKa = 4.81NPTEE465 pKa = 4.33TPSDD469 pKa = 3.34NCTINVTVTDD479 pKa = 3.94DD480 pKa = 3.86DD481 pKa = 4.97YY482 pKa = 12.25GVGTANTKK490 pKa = 8.57VTVNNTKK497 pKa = 10.42PMVEE501 pKa = 4.16APFISLQQNSEE512 pKa = 4.3FILPVVHH519 pKa = 5.85EE520 pKa = 4.29TSFVGTFADD529 pKa = 4.09VGTQDD534 pKa = 2.75THH536 pKa = 4.44TAEE539 pKa = 4.37WNWDD543 pKa = 3.58DD544 pKa = 5.28SSTSDD549 pKa = 3.58GTVTEE554 pKa = 4.47SNGLGKK560 pKa = 10.65VSGSHH565 pKa = 5.41TFSSVGEE572 pKa = 3.95YY573 pKa = 10.32TITLTVTDD581 pKa = 5.44DD582 pKa = 3.45DD583 pKa = 4.36TGADD587 pKa = 3.45SSTKK591 pKa = 9.5TICVVDD597 pKa = 3.62AAEE600 pKa = 5.05ALTITNSYY608 pKa = 9.45IQGLSDD614 pKa = 4.74DD615 pKa = 4.29AFKK618 pKa = 11.35SKK620 pKa = 10.95ADD622 pKa = 3.57QRR624 pKa = 11.84KK625 pKa = 8.79KK626 pKa = 10.99AFDD629 pKa = 3.05KK630 pKa = 10.57MFNVLQDD637 pKa = 3.6RR638 pKa = 11.84LADD641 pKa = 3.37EE642 pKa = 5.52HH643 pKa = 6.63YY644 pKa = 10.64RR645 pKa = 11.84AMIQIMRR652 pKa = 11.84NNIRR656 pKa = 11.84AKK658 pKa = 10.76ADD660 pKa = 3.23GSVDD664 pKa = 3.3GKK666 pKa = 11.47VNNDD670 pKa = 3.72WIIDD674 pKa = 3.81TVAQEE679 pKa = 4.43HH680 pKa = 5.84VCQKK684 pKa = 10.11IDD686 pKa = 4.41DD687 pKa = 3.69ITRR690 pKa = 11.84YY691 pKa = 10.19LEE693 pKa = 3.91YY694 pKa = 10.48LIKK697 pKa = 10.73SS698 pKa = 3.68
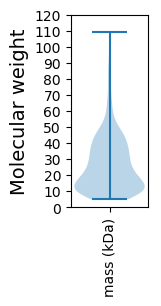
Molecular weight: 77.6 kDa
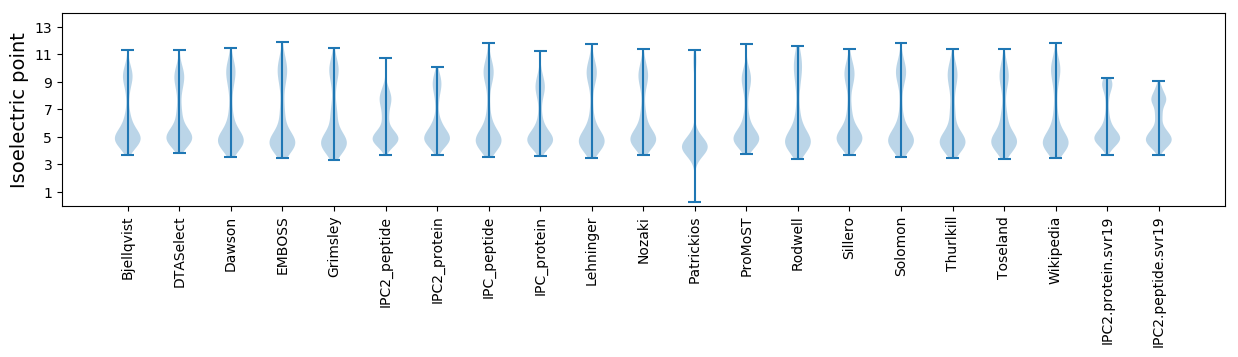
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133VLD5|A0A133VLD5_9EURY Uncharacterized protein OS=candidate division MSBL1 archaeon SCGC-AAA382C18 OX=1698281 GN=AKJ52_00675 PE=4 SV=1
MM1 pKa = 7.07YY2 pKa = 10.71VLALVLSLFEE12 pKa = 4.15VGVGNKK18 pKa = 9.36SFPVFDD24 pKa = 3.87VSPKK28 pKa = 9.16YY29 pKa = 10.72VGWLLRR35 pKa = 11.84IFLPKK40 pKa = 10.11NVSLMRR46 pKa = 11.84ILSKK50 pKa = 10.64LRR52 pKa = 11.84SARR55 pKa = 11.84DD56 pKa = 3.23FRR58 pKa = 11.84GEE60 pKa = 3.72LRR62 pKa = 11.84GVSEE66 pKa = 4.62IGPPAEE72 pKa = 4.5GKK74 pKa = 10.18RR75 pKa = 11.84SPRR78 pKa = 11.84GLFRR82 pKa = 11.84VLGRR86 pKa = 11.84AFGVAKK92 pKa = 10.68QNDD95 pKa = 5.05LILMVV100 pKa = 4.29
MM1 pKa = 7.07YY2 pKa = 10.71VLALVLSLFEE12 pKa = 4.15VGVGNKK18 pKa = 9.36SFPVFDD24 pKa = 3.87VSPKK28 pKa = 9.16YY29 pKa = 10.72VGWLLRR35 pKa = 11.84IFLPKK40 pKa = 10.11NVSLMRR46 pKa = 11.84ILSKK50 pKa = 10.64LRR52 pKa = 11.84SARR55 pKa = 11.84DD56 pKa = 3.23FRR58 pKa = 11.84GEE60 pKa = 3.72LRR62 pKa = 11.84GVSEE66 pKa = 4.62IGPPAEE72 pKa = 4.5GKK74 pKa = 10.18RR75 pKa = 11.84SPRR78 pKa = 11.84GLFRR82 pKa = 11.84VLGRR86 pKa = 11.84AFGVAKK92 pKa = 10.68QNDD95 pKa = 5.05LILMVV100 pKa = 4.29
Molecular weight: 11.16 kDa
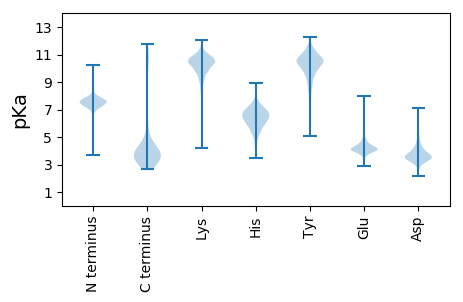
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
131962 |
43 |
955 |
239.5 |
27.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.514 ± 0.103 | 0.907 ± 0.039 |
6.081 ± 0.107 | 10.054 ± 0.212 |
4.075 ± 0.092 | 7.088 ± 0.114 |
1.672 ± 0.044 | 7.529 ± 0.12 |
7.91 ± 0.127 | 9.276 ± 0.139 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.047 | 4.135 ± 0.077 |
3.91 ± 0.071 | 2.302 ± 0.049 |
5.173 ± 0.094 | 6.616 ± 0.09 |
4.721 ± 0.073 | 6.56 ± 0.096 |
1.09 ± 0.038 | 3.152 ± 0.062 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
