
Hamiltosporidium magnivora
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Pansporoblastina; Dubosqiidae; Hamiltosporidium
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
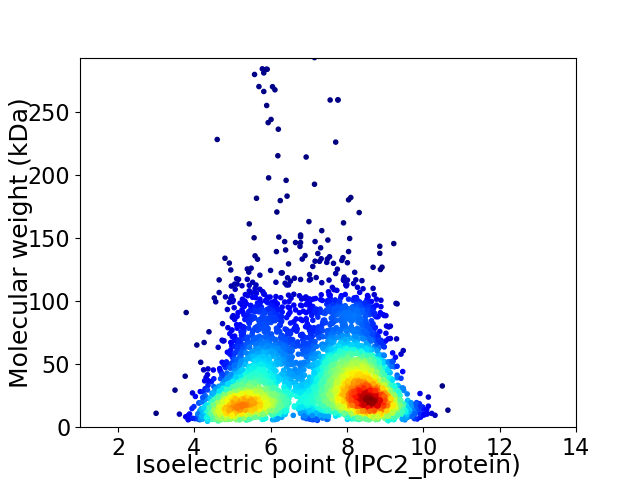
Virtual 2D-PAGE plot for 4459 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
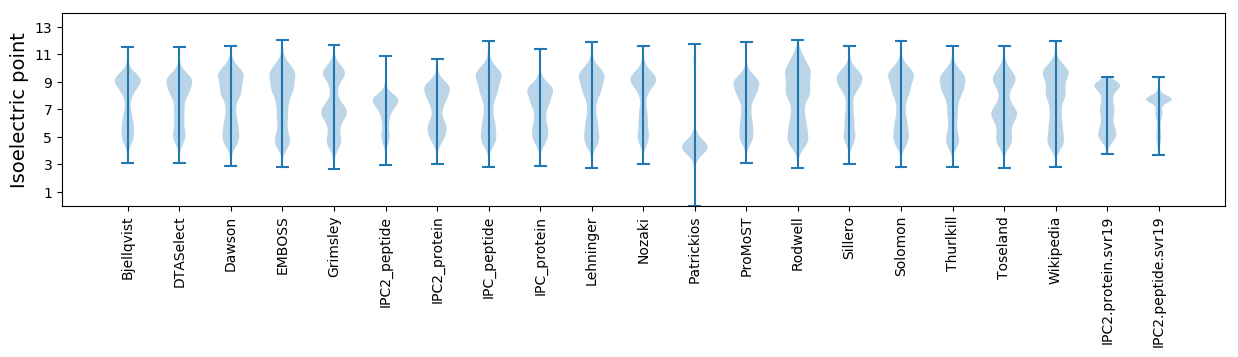
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q9LGS8|A0A4Q9LGS8_9MICR SWI/SNF2-like chromatin-remodeling complex ATPase OS=Hamiltosporidium magnivora OX=148818 GN=CWI36_0319p0020 PE=4 SV=1
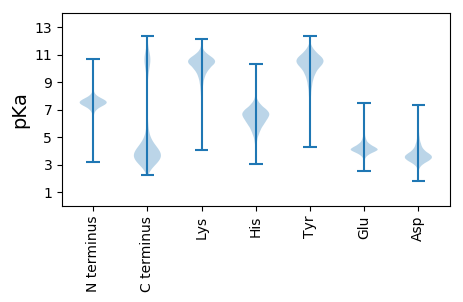
MM1 pKa = 7.18TMIIIVVTNGLNGDD15 pKa = 4.14DD16 pKa = 3.8YY17 pKa = 11.9LLFGMWKK24 pKa = 10.29VKK26 pKa = 9.5MSMMANEE33 pKa = 4.81SNDD36 pKa = 3.32KK37 pKa = 10.83DD38 pKa = 3.64NHH40 pKa = 5.57EE41 pKa = 4.62SCDD44 pKa = 3.52GDD46 pKa = 3.92VGVGDD51 pKa = 5.22VDD53 pKa = 4.09GCDD56 pKa = 4.79DD57 pKa = 5.09KK58 pKa = 11.82DD59 pKa = 4.07DD60 pKa = 4.64HH61 pKa = 7.27DD62 pKa = 4.44SCEE65 pKa = 4.16GATEE69 pKa = 4.67GEE71 pKa = 4.34DD72 pKa = 3.39EE73 pKa = 5.49SDD75 pKa = 3.23IVWYY79 pKa = 9.02TEE81 pKa = 4.53YY82 pKa = 11.32DD83 pKa = 3.95DD84 pKa = 6.18DD85 pKa = 5.7DD86 pKa = 5.32CGYY89 pKa = 11.28DD90 pKa = 3.52DD91 pKa = 6.04GNVEE95 pKa = 4.48GDD97 pKa = 4.09DD98 pKa = 4.38HH99 pKa = 7.45DD100 pKa = 4.68SCDD103 pKa = 5.06GEE105 pKa = 5.5GEE107 pKa = 4.52DD108 pKa = 5.03DD109 pKa = 3.62TDD111 pKa = 3.37GTRR114 pKa = 11.84DD115 pKa = 3.24IKK117 pKa = 11.14YY118 pKa = 10.83DD119 pKa = 3.64SDD121 pKa = 2.88NWGYY125 pKa = 11.33DD126 pKa = 3.05EE127 pKa = 6.4GSDD130 pKa = 5.22DD131 pKa = 4.97RR132 pKa = 11.84DD133 pKa = 3.61DD134 pKa = 5.61HH135 pKa = 6.55EE136 pKa = 5.29SCDD139 pKa = 3.78GATEE143 pKa = 4.27GVDD146 pKa = 3.17GSEE149 pKa = 4.2GIEE152 pKa = 3.76RR153 pKa = 11.84TEE155 pKa = 5.11RR156 pKa = 11.84DD157 pKa = 3.56NDD159 pKa = 3.46DD160 pKa = 4.13CAYY163 pKa = 10.48EE164 pKa = 4.86DD165 pKa = 5.61DD166 pKa = 5.45NDD168 pKa = 5.41DD169 pKa = 4.56GDD171 pKa = 4.54DD172 pKa = 4.04QEE174 pKa = 5.35SCDD177 pKa = 3.7GFGDD181 pKa = 3.65EE182 pKa = 5.18VYY184 pKa = 9.03TLEE187 pKa = 5.41VIFEE191 pKa = 4.18SDD193 pKa = 4.36DD194 pKa = 4.21DD195 pKa = 4.18DD196 pKa = 4.15VVYY199 pKa = 11.41NNGEE203 pKa = 4.33DD204 pKa = 3.25NWEE207 pKa = 4.08GSVVCNDD214 pKa = 3.17KK215 pKa = 11.37KK216 pKa = 10.99EE217 pKa = 4.39DD218 pKa = 3.68GVGIDD223 pKa = 4.48GNDD226 pKa = 3.37SGEE229 pKa = 4.13GNIEE233 pKa = 4.24NISEE237 pKa = 4.24GEE239 pKa = 3.79IGEE242 pKa = 4.19QEE244 pKa = 4.18DD245 pKa = 4.24GKK247 pKa = 10.3VQHH250 pKa = 6.81EE251 pKa = 4.61GTVVIRR257 pKa = 11.84EE258 pKa = 4.05RR259 pKa = 11.84GGDD262 pKa = 3.62RR263 pKa = 11.84FWYY266 pKa = 10.54GDD268 pKa = 3.4FGSDD272 pKa = 2.46EE273 pKa = 4.43KK274 pKa = 11.68YY275 pKa = 10.97SVFEE279 pKa = 4.54DD280 pKa = 3.52LRR282 pKa = 11.84QEE284 pKa = 3.78EE285 pKa = 5.12DD286 pKa = 2.71IWDD289 pKa = 3.83VAEE292 pKa = 5.03LLLKK296 pKa = 10.83KK297 pKa = 10.68LGFAVNYY304 pKa = 10.53AYY306 pKa = 10.34GKK308 pKa = 8.96KK309 pKa = 10.36LKK311 pKa = 10.65VLMVGIGGNSGVVSIKK327 pKa = 9.54TEE329 pKa = 3.91EE330 pKa = 3.82YY331 pKa = 10.4KK332 pKa = 10.92YY333 pKa = 11.1LRR335 pKa = 11.84DD336 pKa = 3.77CDD338 pKa = 3.55ILEE341 pKa = 5.64DD342 pKa = 3.11IWEE345 pKa = 4.63DD346 pKa = 3.75FNSEE350 pKa = 4.57RR351 pKa = 11.84VSDD354 pKa = 4.14CNNGVFEE361 pKa = 4.92NKK363 pKa = 9.96NSGNFEE369 pKa = 4.16TSDD372 pKa = 3.4EE373 pKa = 4.49RR374 pKa = 11.84GDD376 pKa = 3.78GFDD379 pKa = 6.06KK380 pKa = 10.45IDD382 pKa = 3.73EE383 pKa = 4.75DD384 pKa = 4.43GKK386 pKa = 10.85CGKK389 pKa = 10.1GSKK392 pKa = 10.14DD393 pKa = 3.95CEE395 pKa = 4.14DD396 pKa = 3.85CKK398 pKa = 11.13GDD400 pKa = 3.62EE401 pKa = 4.15VDD403 pKa = 3.68KK404 pKa = 11.38HH405 pKa = 7.22GEE407 pKa = 3.81TRR409 pKa = 11.84KK410 pKa = 10.02CGKK413 pKa = 9.35VGKK416 pKa = 9.58FGKK419 pKa = 10.04VVEE422 pKa = 4.15RR423 pKa = 11.84LEE425 pKa = 5.44RR426 pKa = 11.84DD427 pKa = 3.47DD428 pKa = 6.54LKK430 pKa = 11.55DD431 pKa = 3.23NWVEE435 pKa = 4.62CGNCGNSVNWINCGEE450 pKa = 4.15WGNGLIVVIGFVEE463 pKa = 4.75IIVLIEE469 pKa = 3.69WSLKK473 pKa = 9.27IGLVIEE479 pKa = 5.12GIGLADD485 pKa = 4.05LLVAIEE491 pKa = 4.72AFDD494 pKa = 3.83LVEE497 pKa = 4.83PIVLFVVIVLVEE509 pKa = 4.31PIVLFEE515 pKa = 3.9VSMLVEE521 pKa = 4.74IIEE524 pKa = 4.57IIGLIVVIMIEE535 pKa = 4.46MIEE538 pKa = 4.27MIEE541 pKa = 4.21VIVVIVLVEE550 pKa = 4.61PIDD553 pKa = 3.91LVLVGVLVAMIVVIMNLVVFNCNDD577 pKa = 2.85VCGFGGKK584 pKa = 9.34SDD586 pKa = 3.72GVSNDD591 pKa = 2.96MSDD594 pKa = 4.33SKK596 pKa = 11.79DD597 pKa = 3.45CGISGEE603 pKa = 4.19LCFVKK608 pKa = 10.5VGKK611 pKa = 10.35CDD613 pKa = 3.29EE614 pKa = 4.54CVNCAEE620 pKa = 4.66CNGIFEE626 pKa = 4.78GVCDD630 pKa = 4.45CDD632 pKa = 4.15VEE634 pKa = 4.48CTYY637 pKa = 11.2RR638 pKa = 11.84KK639 pKa = 10.01LDD641 pKa = 3.96LYY643 pKa = 10.38CGKK646 pKa = 10.61RR647 pKa = 11.84GGHH650 pKa = 5.84CSKK653 pKa = 11.26CEE655 pKa = 3.97GFDD658 pKa = 4.22GKK660 pKa = 10.72CGCNGNSGKK669 pKa = 10.03IGRR672 pKa = 11.84FVTCGEE678 pKa = 3.87LDD680 pKa = 3.6GRR682 pKa = 11.84CDD684 pKa = 4.67SGSCDD689 pKa = 3.23NCNDD693 pKa = 3.28FPEE696 pKa = 4.65TFGEE700 pKa = 4.29CSEE703 pKa = 4.39SCEE706 pKa = 4.28EE707 pKa = 3.99ASGMDD712 pKa = 4.32TLSAWNTVILFASPGSDD729 pKa = 2.74VVTFAPVGGIGGLGFGLFVGGLFVGGVFVGGVFAGGVFAGGVFAGGVFGGGVFAGGVFVGGVFVGGVFVGGVFAGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVV851 pKa = 2.97
MM1 pKa = 7.18TMIIIVVTNGLNGDD15 pKa = 4.14DD16 pKa = 3.8YY17 pKa = 11.9LLFGMWKK24 pKa = 10.29VKK26 pKa = 9.5MSMMANEE33 pKa = 4.81SNDD36 pKa = 3.32KK37 pKa = 10.83DD38 pKa = 3.64NHH40 pKa = 5.57EE41 pKa = 4.62SCDD44 pKa = 3.52GDD46 pKa = 3.92VGVGDD51 pKa = 5.22VDD53 pKa = 4.09GCDD56 pKa = 4.79DD57 pKa = 5.09KK58 pKa = 11.82DD59 pKa = 4.07DD60 pKa = 4.64HH61 pKa = 7.27DD62 pKa = 4.44SCEE65 pKa = 4.16GATEE69 pKa = 4.67GEE71 pKa = 4.34DD72 pKa = 3.39EE73 pKa = 5.49SDD75 pKa = 3.23IVWYY79 pKa = 9.02TEE81 pKa = 4.53YY82 pKa = 11.32DD83 pKa = 3.95DD84 pKa = 6.18DD85 pKa = 5.7DD86 pKa = 5.32CGYY89 pKa = 11.28DD90 pKa = 3.52DD91 pKa = 6.04GNVEE95 pKa = 4.48GDD97 pKa = 4.09DD98 pKa = 4.38HH99 pKa = 7.45DD100 pKa = 4.68SCDD103 pKa = 5.06GEE105 pKa = 5.5GEE107 pKa = 4.52DD108 pKa = 5.03DD109 pKa = 3.62TDD111 pKa = 3.37GTRR114 pKa = 11.84DD115 pKa = 3.24IKK117 pKa = 11.14YY118 pKa = 10.83DD119 pKa = 3.64SDD121 pKa = 2.88NWGYY125 pKa = 11.33DD126 pKa = 3.05EE127 pKa = 6.4GSDD130 pKa = 5.22DD131 pKa = 4.97RR132 pKa = 11.84DD133 pKa = 3.61DD134 pKa = 5.61HH135 pKa = 6.55EE136 pKa = 5.29SCDD139 pKa = 3.78GATEE143 pKa = 4.27GVDD146 pKa = 3.17GSEE149 pKa = 4.2GIEE152 pKa = 3.76RR153 pKa = 11.84TEE155 pKa = 5.11RR156 pKa = 11.84DD157 pKa = 3.56NDD159 pKa = 3.46DD160 pKa = 4.13CAYY163 pKa = 10.48EE164 pKa = 4.86DD165 pKa = 5.61DD166 pKa = 5.45NDD168 pKa = 5.41DD169 pKa = 4.56GDD171 pKa = 4.54DD172 pKa = 4.04QEE174 pKa = 5.35SCDD177 pKa = 3.7GFGDD181 pKa = 3.65EE182 pKa = 5.18VYY184 pKa = 9.03TLEE187 pKa = 5.41VIFEE191 pKa = 4.18SDD193 pKa = 4.36DD194 pKa = 4.21DD195 pKa = 4.18DD196 pKa = 4.15VVYY199 pKa = 11.41NNGEE203 pKa = 4.33DD204 pKa = 3.25NWEE207 pKa = 4.08GSVVCNDD214 pKa = 3.17KK215 pKa = 11.37KK216 pKa = 10.99EE217 pKa = 4.39DD218 pKa = 3.68GVGIDD223 pKa = 4.48GNDD226 pKa = 3.37SGEE229 pKa = 4.13GNIEE233 pKa = 4.24NISEE237 pKa = 4.24GEE239 pKa = 3.79IGEE242 pKa = 4.19QEE244 pKa = 4.18DD245 pKa = 4.24GKK247 pKa = 10.3VQHH250 pKa = 6.81EE251 pKa = 4.61GTVVIRR257 pKa = 11.84EE258 pKa = 4.05RR259 pKa = 11.84GGDD262 pKa = 3.62RR263 pKa = 11.84FWYY266 pKa = 10.54GDD268 pKa = 3.4FGSDD272 pKa = 2.46EE273 pKa = 4.43KK274 pKa = 11.68YY275 pKa = 10.97SVFEE279 pKa = 4.54DD280 pKa = 3.52LRR282 pKa = 11.84QEE284 pKa = 3.78EE285 pKa = 5.12DD286 pKa = 2.71IWDD289 pKa = 3.83VAEE292 pKa = 5.03LLLKK296 pKa = 10.83KK297 pKa = 10.68LGFAVNYY304 pKa = 10.53AYY306 pKa = 10.34GKK308 pKa = 8.96KK309 pKa = 10.36LKK311 pKa = 10.65VLMVGIGGNSGVVSIKK327 pKa = 9.54TEE329 pKa = 3.91EE330 pKa = 3.82YY331 pKa = 10.4KK332 pKa = 10.92YY333 pKa = 11.1LRR335 pKa = 11.84DD336 pKa = 3.77CDD338 pKa = 3.55ILEE341 pKa = 5.64DD342 pKa = 3.11IWEE345 pKa = 4.63DD346 pKa = 3.75FNSEE350 pKa = 4.57RR351 pKa = 11.84VSDD354 pKa = 4.14CNNGVFEE361 pKa = 4.92NKK363 pKa = 9.96NSGNFEE369 pKa = 4.16TSDD372 pKa = 3.4EE373 pKa = 4.49RR374 pKa = 11.84GDD376 pKa = 3.78GFDD379 pKa = 6.06KK380 pKa = 10.45IDD382 pKa = 3.73EE383 pKa = 4.75DD384 pKa = 4.43GKK386 pKa = 10.85CGKK389 pKa = 10.1GSKK392 pKa = 10.14DD393 pKa = 3.95CEE395 pKa = 4.14DD396 pKa = 3.85CKK398 pKa = 11.13GDD400 pKa = 3.62EE401 pKa = 4.15VDD403 pKa = 3.68KK404 pKa = 11.38HH405 pKa = 7.22GEE407 pKa = 3.81TRR409 pKa = 11.84KK410 pKa = 10.02CGKK413 pKa = 9.35VGKK416 pKa = 9.58FGKK419 pKa = 10.04VVEE422 pKa = 4.15RR423 pKa = 11.84LEE425 pKa = 5.44RR426 pKa = 11.84DD427 pKa = 3.47DD428 pKa = 6.54LKK430 pKa = 11.55DD431 pKa = 3.23NWVEE435 pKa = 4.62CGNCGNSVNWINCGEE450 pKa = 4.15WGNGLIVVIGFVEE463 pKa = 4.75IIVLIEE469 pKa = 3.69WSLKK473 pKa = 9.27IGLVIEE479 pKa = 5.12GIGLADD485 pKa = 4.05LLVAIEE491 pKa = 4.72AFDD494 pKa = 3.83LVEE497 pKa = 4.83PIVLFVVIVLVEE509 pKa = 4.31PIVLFEE515 pKa = 3.9VSMLVEE521 pKa = 4.74IIEE524 pKa = 4.57IIGLIVVIMIEE535 pKa = 4.46MIEE538 pKa = 4.27MIEE541 pKa = 4.21VIVVIVLVEE550 pKa = 4.61PIDD553 pKa = 3.91LVLVGVLVAMIVVIMNLVVFNCNDD577 pKa = 2.85VCGFGGKK584 pKa = 9.34SDD586 pKa = 3.72GVSNDD591 pKa = 2.96MSDD594 pKa = 4.33SKK596 pKa = 11.79DD597 pKa = 3.45CGISGEE603 pKa = 4.19LCFVKK608 pKa = 10.5VGKK611 pKa = 10.35CDD613 pKa = 3.29EE614 pKa = 4.54CVNCAEE620 pKa = 4.66CNGIFEE626 pKa = 4.78GVCDD630 pKa = 4.45CDD632 pKa = 4.15VEE634 pKa = 4.48CTYY637 pKa = 11.2RR638 pKa = 11.84KK639 pKa = 10.01LDD641 pKa = 3.96LYY643 pKa = 10.38CGKK646 pKa = 10.61RR647 pKa = 11.84GGHH650 pKa = 5.84CSKK653 pKa = 11.26CEE655 pKa = 3.97GFDD658 pKa = 4.22GKK660 pKa = 10.72CGCNGNSGKK669 pKa = 10.03IGRR672 pKa = 11.84FVTCGEE678 pKa = 3.87LDD680 pKa = 3.6GRR682 pKa = 11.84CDD684 pKa = 4.67SGSCDD689 pKa = 3.23NCNDD693 pKa = 3.28FPEE696 pKa = 4.65TFGEE700 pKa = 4.29CSEE703 pKa = 4.39SCEE706 pKa = 4.28EE707 pKa = 3.99ASGMDD712 pKa = 4.32TLSAWNTVILFASPGSDD729 pKa = 2.74VVTFAPVGGIGGLGFGLFVGGLFVGGVFVGGVFAGGVFAGGVFAGGVFGGGVFAGGVFVGGVFVGGVFVGGVFAGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVFVGGVV851 pKa = 2.97
Molecular weight: 90.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q9LMK5|A0A4Q9LMK5_9MICR Uncharacterized protein (Fragment) OS=Hamiltosporidium magnivora OX=148818 GN=CWI36_0022p0050 PE=4 SV=1
MM1 pKa = 7.84IILNILSKK9 pKa = 10.43ISRR12 pKa = 11.84VCASFSSRR20 pKa = 11.84SYY22 pKa = 10.08KK23 pKa = 10.38SSSSRR28 pKa = 11.84VSSHH32 pKa = 6.49SGGGSGKK39 pKa = 8.88TSLSHH44 pKa = 6.51SGYY47 pKa = 9.64SSSGGGSDD55 pKa = 3.68GSSSSRR61 pKa = 11.84GSSSGGSSSSGGGSGGSSSSSGSSSSGGSSSSGGSSGSGGSSGSGGGSGSGGGSGSRR118 pKa = 11.84GGSGSRR124 pKa = 11.84GGSGSGGGSGSGGGSGGSTGSSSSQGSSNGKK155 pKa = 9.17CCPCPSS161 pKa = 3.45
MM1 pKa = 7.84IILNILSKK9 pKa = 10.43ISRR12 pKa = 11.84VCASFSSRR20 pKa = 11.84SYY22 pKa = 10.08KK23 pKa = 10.38SSSSRR28 pKa = 11.84VSSHH32 pKa = 6.49SGGGSGKK39 pKa = 8.88TSLSHH44 pKa = 6.51SGYY47 pKa = 9.64SSSGGGSDD55 pKa = 3.68GSSSSRR61 pKa = 11.84GSSSGGSSSSGGGSGGSSSSSGSSSSGGSSSSGGSSGSGGSSGSGGGSGSGGGSGSRR118 pKa = 11.84GGSGSRR124 pKa = 11.84GGSGSGGGSGSGGGSGGSTGSSSSQGSSNGKK155 pKa = 9.17CCPCPSS161 pKa = 3.45
Molecular weight: 13.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1572801 |
41 |
2447 |
352.7 |
41.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.028 | 1.959 ± 0.018 |
5.207 ± 0.026 | 7.493 ± 0.04 |
6.36 ± 0.043 | 3.504 ± 0.043 |
1.651 ± 0.015 | 9.96 ± 0.037 |
9.942 ± 0.05 | 9.582 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.94 ± 0.014 | 8.785 ± 0.047 |
2.536 ± 0.025 | 2.487 ± 0.019 |
3.534 ± 0.025 | 7.864 ± 0.031 |
4.988 ± 0.035 | 4.596 ± 0.03 |
0.489 ± 0.007 | 4.648 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
