
Capybara microvirus Cap1_SP_141
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.21
Get precalculated fractions of proteins
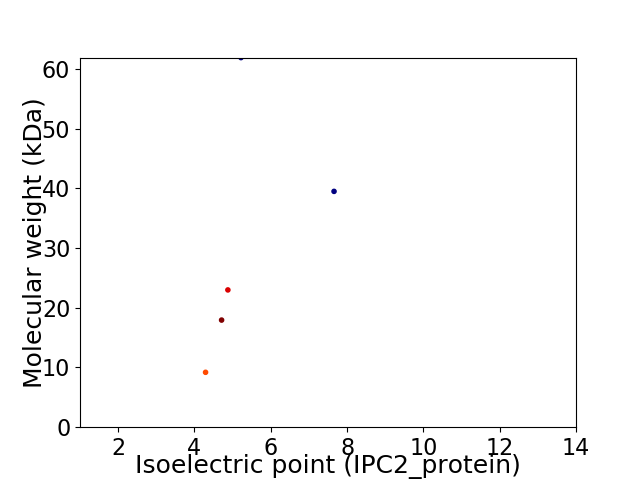
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
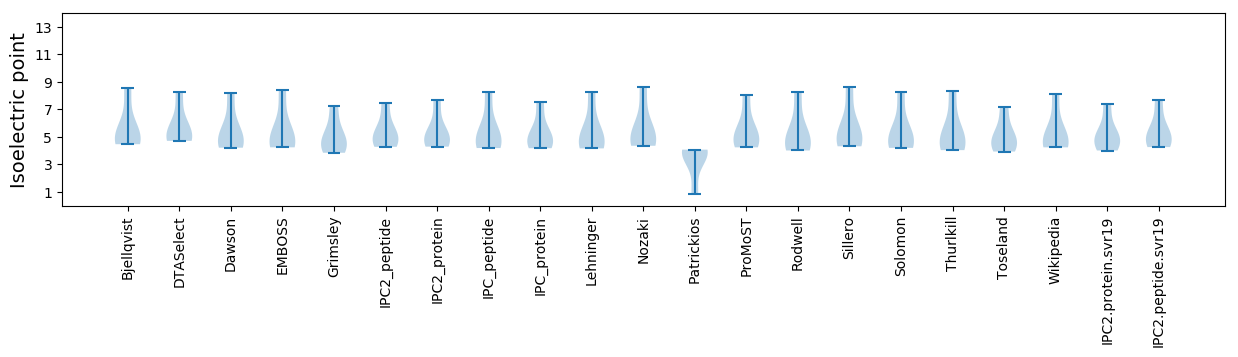
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5E7|A0A4P8W5E7_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_141 OX=2585387 PE=4 SV=1
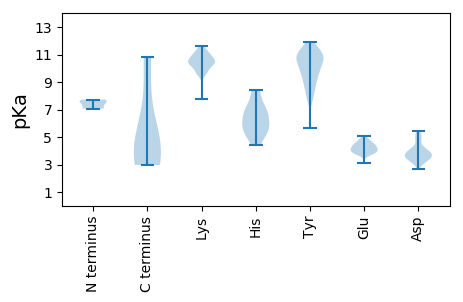
MM1 pKa = 7.52IKK3 pKa = 10.17SIYY6 pKa = 9.48CIRR9 pKa = 11.84DD10 pKa = 3.23VAIGYY15 pKa = 8.38CDD17 pKa = 4.02PFCALNDD24 pKa = 3.57KK25 pKa = 10.61VAIRR29 pKa = 11.84DD30 pKa = 3.45FRR32 pKa = 11.84NAINLPNTPYY42 pKa = 11.17NLNPKK47 pKa = 10.02DD48 pKa = 3.93YY49 pKa = 11.32DD50 pKa = 4.05LMCVASIDD58 pKa = 4.15LDD60 pKa = 3.79SGTVVAQQPQLVVSGISLVGVNNDD84 pKa = 2.97
MM1 pKa = 7.52IKK3 pKa = 10.17SIYY6 pKa = 9.48CIRR9 pKa = 11.84DD10 pKa = 3.23VAIGYY15 pKa = 8.38CDD17 pKa = 4.02PFCALNDD24 pKa = 3.57KK25 pKa = 10.61VAIRR29 pKa = 11.84DD30 pKa = 3.45FRR32 pKa = 11.84NAINLPNTPYY42 pKa = 11.17NLNPKK47 pKa = 10.02DD48 pKa = 3.93YY49 pKa = 11.32DD50 pKa = 4.05LMCVASIDD58 pKa = 4.15LDD60 pKa = 3.79SGTVVAQQPQLVVSGISLVGVNNDD84 pKa = 2.97
Molecular weight: 9.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7Y7|A0A4P8W7Y7_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_141 OX=2585387 PE=4 SV=1
MM1 pKa = 7.07CTSPKK6 pKa = 10.24KK7 pKa = 10.34AFCVGLTKK15 pKa = 10.51NGKK18 pKa = 7.75PDD20 pKa = 3.69YY21 pKa = 10.98VIAPYY26 pKa = 9.9QCHH29 pKa = 5.48HH30 pKa = 6.1VEE32 pKa = 4.25KK33 pKa = 10.51IGDD36 pKa = 3.59KK37 pKa = 10.53FEE39 pKa = 4.45RR40 pKa = 11.84VYY42 pKa = 11.2SSAMSPRR49 pKa = 11.84ADD51 pKa = 2.92LVVSTFIEE59 pKa = 4.79IPCGRR64 pKa = 11.84CLDD67 pKa = 3.88CRR69 pKa = 11.84LSYY72 pKa = 10.57SRR74 pKa = 11.84SWADD78 pKa = 3.39RR79 pKa = 11.84LVRR82 pKa = 11.84EE83 pKa = 4.64SKK85 pKa = 9.91TSVSSLFVTFTYY97 pKa = 10.96DD98 pKa = 4.22DD99 pKa = 3.6DD100 pKa = 4.06HH101 pKa = 8.45LPYY104 pKa = 9.98HH105 pKa = 7.36DD106 pKa = 4.49YY107 pKa = 11.52VNSEE111 pKa = 3.96SGEE114 pKa = 4.2LVSKK118 pKa = 10.14PCLVKK123 pKa = 10.33KK124 pKa = 9.89HH125 pKa = 6.11CQDD128 pKa = 2.89FFKK131 pKa = 10.81RR132 pKa = 11.84LRR134 pKa = 11.84KK135 pKa = 10.2YY136 pKa = 10.56FDD138 pKa = 4.02GYY140 pKa = 10.05NIRR143 pKa = 11.84YY144 pKa = 8.77FIAGEE149 pKa = 4.01YY150 pKa = 10.57GEE152 pKa = 4.63TSARR156 pKa = 11.84PHH158 pKa = 4.91YY159 pKa = 10.42HH160 pKa = 5.58AVIYY164 pKa = 9.32NLPLDD169 pKa = 4.07KK170 pKa = 11.05LDD172 pKa = 3.53LQFYY176 pKa = 7.8KK177 pKa = 10.98TNFNGDD183 pKa = 3.75LYY185 pKa = 9.39YY186 pKa = 10.47TSPVLEE192 pKa = 4.93SIWTHH197 pKa = 5.76GYY199 pKa = 9.81VVLSDD204 pKa = 4.71FSWQTGAYY212 pKa = 5.63TARR215 pKa = 11.84YY216 pKa = 8.46ILKK219 pKa = 9.72KK220 pKa = 10.52QFGKK224 pKa = 10.76ASDD227 pKa = 3.65QFKK230 pKa = 9.69LTGFPQEE237 pKa = 3.96FVLMSRR243 pKa = 11.84KK244 pKa = 9.48PGLGRR249 pKa = 11.84QYY251 pKa = 11.04FNEE254 pKa = 4.58NIDD257 pKa = 4.71DD258 pKa = 5.19LIAYY262 pKa = 9.61DD263 pKa = 5.12GDD265 pKa = 3.47ICRR268 pKa = 11.84PGQKK272 pKa = 9.64SKK274 pKa = 10.42PGRR277 pKa = 11.84YY278 pKa = 8.72FDD280 pKa = 5.41SLIEE284 pKa = 4.47DD285 pKa = 4.68ADD287 pKa = 3.78PEE289 pKa = 4.41KK290 pKa = 11.41YY291 pKa = 10.07KK292 pKa = 10.66DD293 pKa = 3.72LKK295 pKa = 11.19NNRR298 pKa = 11.84QTVRR302 pKa = 11.84DD303 pKa = 3.61LFIAGKK309 pKa = 9.53LAQTDD314 pKa = 3.49KK315 pKa = 11.45SYY317 pKa = 11.9LDD319 pKa = 3.71LLSVEE324 pKa = 4.13DD325 pKa = 3.84HH326 pKa = 7.03YY327 pKa = 10.98YY328 pKa = 7.85TQRR331 pKa = 11.84ALHH334 pKa = 6.77LKK336 pKa = 10.51RR337 pKa = 11.84KK338 pKa = 9.85DD339 pKa = 3.31LL340 pKa = 4.04
MM1 pKa = 7.07CTSPKK6 pKa = 10.24KK7 pKa = 10.34AFCVGLTKK15 pKa = 10.51NGKK18 pKa = 7.75PDD20 pKa = 3.69YY21 pKa = 10.98VIAPYY26 pKa = 9.9QCHH29 pKa = 5.48HH30 pKa = 6.1VEE32 pKa = 4.25KK33 pKa = 10.51IGDD36 pKa = 3.59KK37 pKa = 10.53FEE39 pKa = 4.45RR40 pKa = 11.84VYY42 pKa = 11.2SSAMSPRR49 pKa = 11.84ADD51 pKa = 2.92LVVSTFIEE59 pKa = 4.79IPCGRR64 pKa = 11.84CLDD67 pKa = 3.88CRR69 pKa = 11.84LSYY72 pKa = 10.57SRR74 pKa = 11.84SWADD78 pKa = 3.39RR79 pKa = 11.84LVRR82 pKa = 11.84EE83 pKa = 4.64SKK85 pKa = 9.91TSVSSLFVTFTYY97 pKa = 10.96DD98 pKa = 4.22DD99 pKa = 3.6DD100 pKa = 4.06HH101 pKa = 8.45LPYY104 pKa = 9.98HH105 pKa = 7.36DD106 pKa = 4.49YY107 pKa = 11.52VNSEE111 pKa = 3.96SGEE114 pKa = 4.2LVSKK118 pKa = 10.14PCLVKK123 pKa = 10.33KK124 pKa = 9.89HH125 pKa = 6.11CQDD128 pKa = 2.89FFKK131 pKa = 10.81RR132 pKa = 11.84LRR134 pKa = 11.84KK135 pKa = 10.2YY136 pKa = 10.56FDD138 pKa = 4.02GYY140 pKa = 10.05NIRR143 pKa = 11.84YY144 pKa = 8.77FIAGEE149 pKa = 4.01YY150 pKa = 10.57GEE152 pKa = 4.63TSARR156 pKa = 11.84PHH158 pKa = 4.91YY159 pKa = 10.42HH160 pKa = 5.58AVIYY164 pKa = 9.32NLPLDD169 pKa = 4.07KK170 pKa = 11.05LDD172 pKa = 3.53LQFYY176 pKa = 7.8KK177 pKa = 10.98TNFNGDD183 pKa = 3.75LYY185 pKa = 9.39YY186 pKa = 10.47TSPVLEE192 pKa = 4.93SIWTHH197 pKa = 5.76GYY199 pKa = 9.81VVLSDD204 pKa = 4.71FSWQTGAYY212 pKa = 5.63TARR215 pKa = 11.84YY216 pKa = 8.46ILKK219 pKa = 9.72KK220 pKa = 10.52QFGKK224 pKa = 10.76ASDD227 pKa = 3.65QFKK230 pKa = 9.69LTGFPQEE237 pKa = 3.96FVLMSRR243 pKa = 11.84KK244 pKa = 9.48PGLGRR249 pKa = 11.84QYY251 pKa = 11.04FNEE254 pKa = 4.58NIDD257 pKa = 4.71DD258 pKa = 5.19LIAYY262 pKa = 9.61DD263 pKa = 5.12GDD265 pKa = 3.47ICRR268 pKa = 11.84PGQKK272 pKa = 9.64SKK274 pKa = 10.42PGRR277 pKa = 11.84YY278 pKa = 8.72FDD280 pKa = 5.41SLIEE284 pKa = 4.47DD285 pKa = 4.68ADD287 pKa = 3.78PEE289 pKa = 4.41KK290 pKa = 11.41YY291 pKa = 10.07KK292 pKa = 10.66DD293 pKa = 3.72LKK295 pKa = 11.19NNRR298 pKa = 11.84QTVRR302 pKa = 11.84DD303 pKa = 3.61LFIAGKK309 pKa = 9.53LAQTDD314 pKa = 3.49KK315 pKa = 11.45SYY317 pKa = 11.9LDD319 pKa = 3.71LLSVEE324 pKa = 4.13DD325 pKa = 3.84HH326 pKa = 7.03YY327 pKa = 10.98YY328 pKa = 7.85TQRR331 pKa = 11.84ALHH334 pKa = 6.77LKK336 pKa = 10.51RR337 pKa = 11.84KK338 pKa = 9.85DD339 pKa = 3.31LL340 pKa = 4.04
Molecular weight: 39.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1362 |
84 |
558 |
272.4 |
30.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.461 ± 0.957 | 1.175 ± 0.601 |
6.902 ± 0.835 | 4.185 ± 0.742 |
4.993 ± 0.691 | 7.489 ± 1.054 |
1.762 ± 0.353 | 4.846 ± 0.624 |
5.507 ± 1.015 | 8.076 ± 0.494 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.129 ± 0.411 | 6.021 ± 0.811 |
5.36 ± 0.257 | 4.038 ± 0.542 |
3.818 ± 0.725 | 8.443 ± 1.235 |
5.8 ± 0.553 | 6.828 ± 0.689 |
1.101 ± 0.233 | 5.066 ± 0.711 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
