
Anabarilius grahami (Kanglang fish) (Barilius grahami)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Otomorpha; Ostariophysi; Otophysi;
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
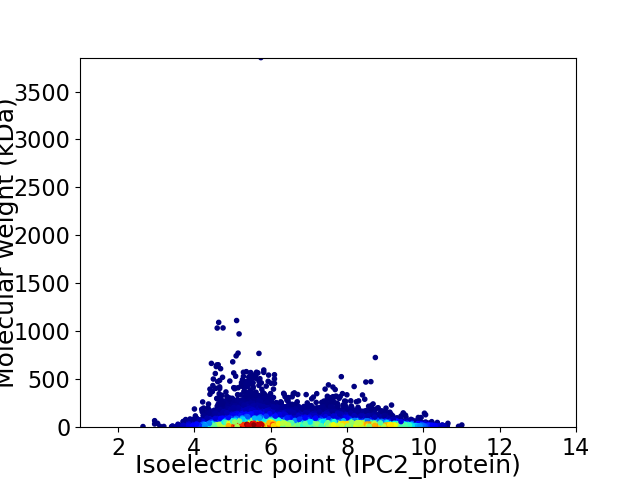
Virtual 2D-PAGE plot for 23663 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
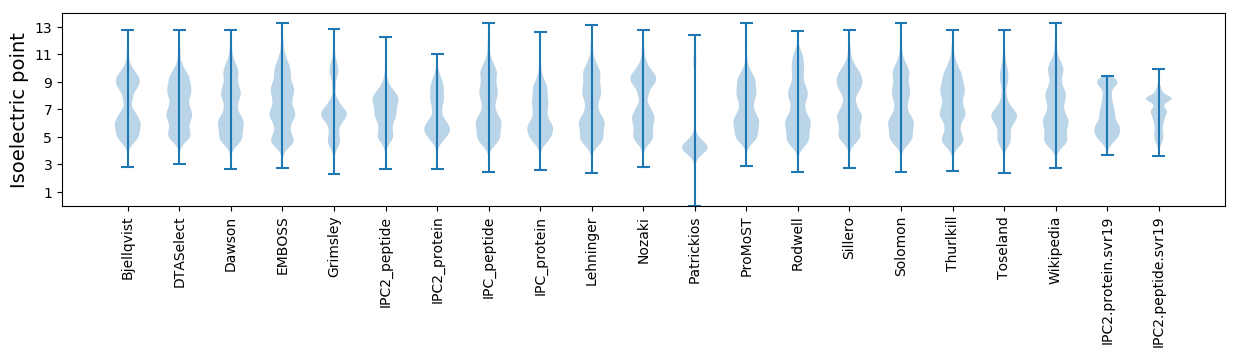
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N0XLN5|A0A3N0XLN5_ANAGA Uncharacterized protein OS=Anabarilius grahami OX=495550 GN=DPX16_2126 PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 4.94RR3 pKa = 11.84NIYY6 pKa = 9.1MLIRR10 pKa = 11.84HH11 pKa = 6.42RR12 pKa = 11.84NGQSTLHH19 pKa = 5.61ATIIDD24 pKa = 3.52GDD26 pKa = 3.82VRR28 pKa = 11.84RR29 pKa = 11.84EE30 pKa = 3.77IPEE33 pKa = 3.8IAVPSHH39 pKa = 7.38DD40 pKa = 3.06EE41 pKa = 3.83VLFNRR46 pKa = 11.84VVQAVCRR53 pKa = 11.84LTRR56 pKa = 11.84YY57 pKa = 9.21PLTPAPDD64 pKa = 3.29RR65 pKa = 11.84QYY67 pKa = 11.89NPLLSMLAWNADD79 pKa = 2.98AWGVSTMTATGDD91 pKa = 3.35EE92 pKa = 4.34SDD94 pKa = 3.09IAFYY98 pKa = 10.0PVYY101 pKa = 10.5NNPPTEE107 pKa = 3.76RR108 pKa = 11.84VEE110 pKa = 4.02RR111 pKa = 11.84FRR113 pKa = 11.84IEE115 pKa = 3.94HH116 pKa = 6.75RR117 pKa = 11.84SVQPRR122 pKa = 11.84PSGEE126 pKa = 3.93SHH128 pKa = 4.69MHH130 pKa = 6.78GSRR133 pKa = 11.84TDD135 pKa = 3.58DD136 pKa = 3.33EE137 pKa = 4.93HH138 pKa = 7.32PEE140 pKa = 4.02PSHH143 pKa = 6.97LDD145 pKa = 3.44NNSEE149 pKa = 3.92IGSDD153 pKa = 3.69FDD155 pKa = 4.97RR156 pKa = 11.84SSEE159 pKa = 4.21SLGSSLIGEE168 pKa = 4.97ASTDD172 pKa = 3.71DD173 pKa = 4.1DD174 pKa = 4.41QSGSSQMDD182 pKa = 3.53NNSDD186 pKa = 3.06SGYY189 pKa = 10.33YY190 pKa = 10.75SNDD193 pKa = 3.05SSDD196 pKa = 3.86SLSSSFNPQASLGEE210 pKa = 3.96TSRR213 pKa = 11.84DD214 pKa = 3.6DD215 pKa = 3.85EE216 pKa = 4.62QPGPSHH222 pKa = 6.93LVNNSKK228 pKa = 9.83IRR230 pKa = 11.84SNSDD234 pKa = 2.97CSINSLSSSCFPPASLRR251 pKa = 11.84EE252 pKa = 3.97TSTDD256 pKa = 3.52DD257 pKa = 3.84DD258 pKa = 5.08LSGSSHH264 pKa = 6.53MDD266 pKa = 3.1NNSEE270 pKa = 4.04IVFDD274 pKa = 4.48SDD276 pKa = 3.79CSSDD280 pKa = 3.22SFYY283 pKa = 11.64SLASLGEE290 pKa = 3.81ISTDD294 pKa = 3.6DD295 pKa = 4.08DD296 pKa = 4.87LSGSSHH302 pKa = 5.41MVNNSEE308 pKa = 4.59IGFDD312 pKa = 3.94SDD314 pKa = 3.93CSSDD318 pKa = 3.22SFYY321 pKa = 11.64SLASLGEE328 pKa = 3.81ISTDD332 pKa = 3.68DD333 pKa = 4.25DD334 pKa = 4.99LSGSPHH340 pKa = 5.64MVNNSEE346 pKa = 4.17IGFDD350 pKa = 3.94SDD352 pKa = 3.93CSSDD356 pKa = 3.22SFYY359 pKa = 11.64SLASLGEE366 pKa = 3.81ISTDD370 pKa = 3.68DD371 pKa = 4.25DD372 pKa = 4.99LSGSPHH378 pKa = 5.64MVNNSEE384 pKa = 4.17IGFDD388 pKa = 3.94SDD390 pKa = 3.93CSSDD394 pKa = 3.22SFYY397 pKa = 11.64SLASLGEE404 pKa = 3.81ISTDD408 pKa = 3.68DD409 pKa = 4.25DD410 pKa = 4.99LSGSPHH416 pKa = 5.64MVNNSEE422 pKa = 4.17IGFDD426 pKa = 3.94SDD428 pKa = 3.93CSSDD432 pKa = 3.22SFYY435 pKa = 11.64SLASLGEE442 pKa = 3.81ISTDD446 pKa = 3.68DD447 pKa = 4.25DD448 pKa = 4.99LSGSPHH454 pKa = 5.64MVNNSEE460 pKa = 4.17IGFDD464 pKa = 3.94SDD466 pKa = 3.93CSSDD470 pKa = 3.22SFYY473 pKa = 11.64SLASLGEE480 pKa = 3.81ISTDD484 pKa = 3.68DD485 pKa = 4.25DD486 pKa = 4.99LSGSPHH492 pKa = 5.64MVNNSEE498 pKa = 4.17IGFDD502 pKa = 3.94SDD504 pKa = 3.93CSSDD508 pKa = 3.22SFYY511 pKa = 11.64SLASLGEE518 pKa = 3.81ISTDD522 pKa = 3.68DD523 pKa = 4.25DD524 pKa = 4.99LSGSPHH530 pKa = 5.64MVNNSEE536 pKa = 4.17IGFDD540 pKa = 3.79SDD542 pKa = 4.03CTLDD546 pKa = 3.94LSS548 pKa = 4.34
MM1 pKa = 7.87DD2 pKa = 4.94RR3 pKa = 11.84NIYY6 pKa = 9.1MLIRR10 pKa = 11.84HH11 pKa = 6.42RR12 pKa = 11.84NGQSTLHH19 pKa = 5.61ATIIDD24 pKa = 3.52GDD26 pKa = 3.82VRR28 pKa = 11.84RR29 pKa = 11.84EE30 pKa = 3.77IPEE33 pKa = 3.8IAVPSHH39 pKa = 7.38DD40 pKa = 3.06EE41 pKa = 3.83VLFNRR46 pKa = 11.84VVQAVCRR53 pKa = 11.84LTRR56 pKa = 11.84YY57 pKa = 9.21PLTPAPDD64 pKa = 3.29RR65 pKa = 11.84QYY67 pKa = 11.89NPLLSMLAWNADD79 pKa = 2.98AWGVSTMTATGDD91 pKa = 3.35EE92 pKa = 4.34SDD94 pKa = 3.09IAFYY98 pKa = 10.0PVYY101 pKa = 10.5NNPPTEE107 pKa = 3.76RR108 pKa = 11.84VEE110 pKa = 4.02RR111 pKa = 11.84FRR113 pKa = 11.84IEE115 pKa = 3.94HH116 pKa = 6.75RR117 pKa = 11.84SVQPRR122 pKa = 11.84PSGEE126 pKa = 3.93SHH128 pKa = 4.69MHH130 pKa = 6.78GSRR133 pKa = 11.84TDD135 pKa = 3.58DD136 pKa = 3.33EE137 pKa = 4.93HH138 pKa = 7.32PEE140 pKa = 4.02PSHH143 pKa = 6.97LDD145 pKa = 3.44NNSEE149 pKa = 3.92IGSDD153 pKa = 3.69FDD155 pKa = 4.97RR156 pKa = 11.84SSEE159 pKa = 4.21SLGSSLIGEE168 pKa = 4.97ASTDD172 pKa = 3.71DD173 pKa = 4.1DD174 pKa = 4.41QSGSSQMDD182 pKa = 3.53NNSDD186 pKa = 3.06SGYY189 pKa = 10.33YY190 pKa = 10.75SNDD193 pKa = 3.05SSDD196 pKa = 3.86SLSSSFNPQASLGEE210 pKa = 3.96TSRR213 pKa = 11.84DD214 pKa = 3.6DD215 pKa = 3.85EE216 pKa = 4.62QPGPSHH222 pKa = 6.93LVNNSKK228 pKa = 9.83IRR230 pKa = 11.84SNSDD234 pKa = 2.97CSINSLSSSCFPPASLRR251 pKa = 11.84EE252 pKa = 3.97TSTDD256 pKa = 3.52DD257 pKa = 3.84DD258 pKa = 5.08LSGSSHH264 pKa = 6.53MDD266 pKa = 3.1NNSEE270 pKa = 4.04IVFDD274 pKa = 4.48SDD276 pKa = 3.79CSSDD280 pKa = 3.22SFYY283 pKa = 11.64SLASLGEE290 pKa = 3.81ISTDD294 pKa = 3.6DD295 pKa = 4.08DD296 pKa = 4.87LSGSSHH302 pKa = 5.41MVNNSEE308 pKa = 4.59IGFDD312 pKa = 3.94SDD314 pKa = 3.93CSSDD318 pKa = 3.22SFYY321 pKa = 11.64SLASLGEE328 pKa = 3.81ISTDD332 pKa = 3.68DD333 pKa = 4.25DD334 pKa = 4.99LSGSPHH340 pKa = 5.64MVNNSEE346 pKa = 4.17IGFDD350 pKa = 3.94SDD352 pKa = 3.93CSSDD356 pKa = 3.22SFYY359 pKa = 11.64SLASLGEE366 pKa = 3.81ISTDD370 pKa = 3.68DD371 pKa = 4.25DD372 pKa = 4.99LSGSPHH378 pKa = 5.64MVNNSEE384 pKa = 4.17IGFDD388 pKa = 3.94SDD390 pKa = 3.93CSSDD394 pKa = 3.22SFYY397 pKa = 11.64SLASLGEE404 pKa = 3.81ISTDD408 pKa = 3.68DD409 pKa = 4.25DD410 pKa = 4.99LSGSPHH416 pKa = 5.64MVNNSEE422 pKa = 4.17IGFDD426 pKa = 3.94SDD428 pKa = 3.93CSSDD432 pKa = 3.22SFYY435 pKa = 11.64SLASLGEE442 pKa = 3.81ISTDD446 pKa = 3.68DD447 pKa = 4.25DD448 pKa = 4.99LSGSPHH454 pKa = 5.64MVNNSEE460 pKa = 4.17IGFDD464 pKa = 3.94SDD466 pKa = 3.93CSSDD470 pKa = 3.22SFYY473 pKa = 11.64SLASLGEE480 pKa = 3.81ISTDD484 pKa = 3.68DD485 pKa = 4.25DD486 pKa = 4.99LSGSPHH492 pKa = 5.64MVNNSEE498 pKa = 4.17IGFDD502 pKa = 3.94SDD504 pKa = 3.93CSSDD508 pKa = 3.22SFYY511 pKa = 11.64SLASLGEE518 pKa = 3.81ISTDD522 pKa = 3.68DD523 pKa = 4.25DD524 pKa = 4.99LSGSPHH530 pKa = 5.64MVNNSEE536 pKa = 4.17IGFDD540 pKa = 3.79SDD542 pKa = 4.03CTLDD546 pKa = 3.94LSS548 pKa = 4.34
Molecular weight: 58.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N0XMM2|A0A3N0XMM2_ANAGA Tyrosine-protein phosphatase non-receptor type 21 OS=Anabarilius grahami OX=495550 GN=DPX16_2139 PE=3 SV=1
MM1 pKa = 7.01VVQVAQSTMVVQVAQGTMAVQVAQGTMAMQVAQGTMVVQVAQGTMAVQVAQGTMAMQVAQGTMVVQLAQGPMVVQVAQRR80 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR107 pKa = 11.84PMVVQVAHH115 pKa = 7.08RR116 pKa = 11.84PMAVQVAHH124 pKa = 7.29RR125 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR152 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR179 pKa = 11.84PMVVQVAHH187 pKa = 6.96RR188 pKa = 11.84PMVVQVAQGTMVMQVAQGAMVMQVAQGAMVVV219 pKa = 3.42
MM1 pKa = 7.01VVQVAQSTMVVQVAQGTMAVQVAQGTMAMQVAQGTMVVQVAQGTMAVQVAQGTMAMQVAQGTMVVQLAQGPMVVQVAQRR80 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR107 pKa = 11.84PMVVQVAHH115 pKa = 7.08RR116 pKa = 11.84PMAVQVAHH124 pKa = 7.29RR125 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR152 pKa = 11.84PMAMQVAQGTMVVQLAQGPMVVQVAQRR179 pKa = 11.84PMVVQVAHH187 pKa = 6.96RR188 pKa = 11.84PMVVQVAQGTMVMQVAQGAMVMQVAQGAMVVV219 pKa = 3.42
Molecular weight: 22.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11186444 |
49 |
34580 |
472.7 |
52.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.37 ± 0.016 | 2.315 ± 0.013 |
5.182 ± 0.012 | 6.988 ± 0.029 |
3.624 ± 0.013 | 5.938 ± 0.022 |
2.69 ± 0.01 | 4.554 ± 0.014 |
5.743 ± 0.026 | 9.432 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.379 ± 0.009 | 3.895 ± 0.012 |
5.598 ± 0.022 | 4.676 ± 0.019 |
5.754 ± 0.016 | 8.87 ± 0.026 |
5.787 ± 0.018 | 6.361 ± 0.019 |
1.172 ± 0.007 | 2.619 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
