
Beihai picobirna-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
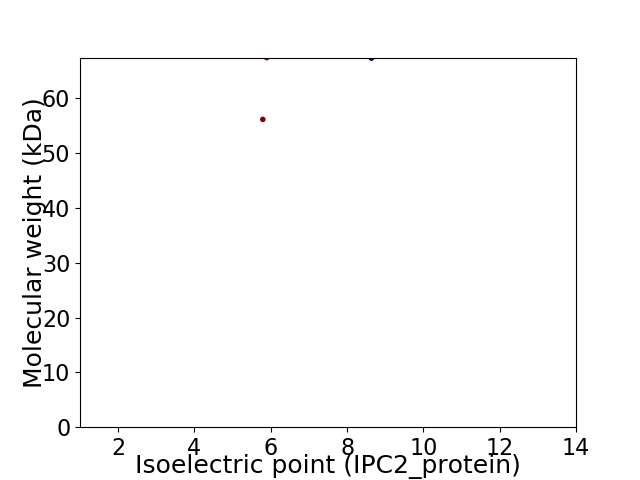
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
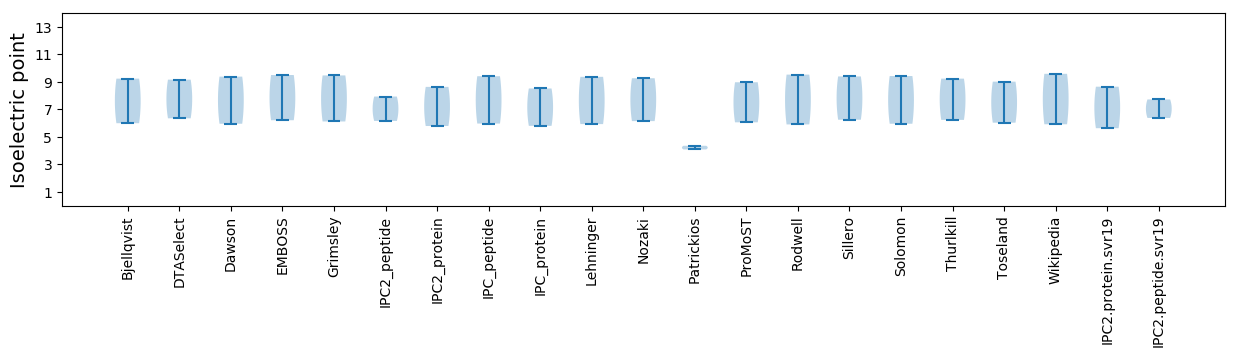
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLD7|A0A1L3KLD7_9VIRU Uncharacterized protein OS=Beihai picobirna-like virus 7 OX=1922524 PE=4 SV=1
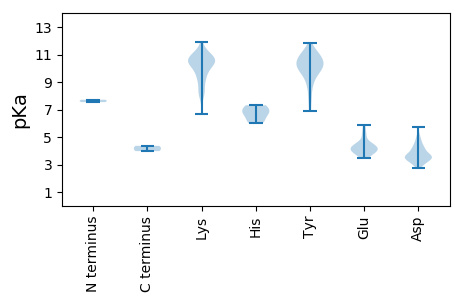
MM1 pKa = 7.54NFQFLDD7 pKa = 3.6TLSVSKK13 pKa = 10.57QVEE16 pKa = 4.11NSVSSNLRR24 pKa = 11.84RR25 pKa = 11.84VVSGSEE31 pKa = 4.12DD32 pKa = 3.6VLTSPIVKK40 pKa = 9.98RR41 pKa = 11.84KK42 pKa = 9.96APEE45 pKa = 5.16DD46 pKa = 3.39ILKK49 pKa = 10.54GWDD52 pKa = 3.53EE53 pKa = 3.93IVKK56 pKa = 10.44EE57 pKa = 4.26NLSAMNADD65 pKa = 4.97LINLEE70 pKa = 4.49EE71 pKa = 4.34SNKK74 pKa = 10.52LKK76 pKa = 10.68FGPRR80 pKa = 11.84SIAAPWGEE88 pKa = 3.97RR89 pKa = 11.84TDD91 pKa = 4.21GVLSYY96 pKa = 10.31FAEE99 pKa = 4.58DD100 pKa = 3.68TLFTPPTVSKK110 pKa = 10.68VPRR113 pKa = 11.84RR114 pKa = 11.84NLRR117 pKa = 11.84PISEE121 pKa = 4.05AKK123 pKa = 10.7ALGFLKK129 pKa = 10.79NSTNSGLPYY138 pKa = 8.56YY139 pKa = 8.87TKK141 pKa = 10.25KK142 pKa = 11.04GKK144 pKa = 10.37VKK146 pKa = 10.28EE147 pKa = 4.5IYY149 pKa = 9.31STMQSPLLSRR159 pKa = 11.84KK160 pKa = 9.55DD161 pKa = 3.36PCVMFTRR168 pKa = 11.84TQEE171 pKa = 3.75QRR173 pKa = 11.84KK174 pKa = 6.66TRR176 pKa = 11.84TVWGYY181 pKa = 10.32PMADD185 pKa = 3.69TLRR188 pKa = 11.84EE189 pKa = 3.85MQYY192 pKa = 10.54YY193 pKa = 9.88EE194 pKa = 5.39PILGYY199 pKa = 7.76QRR201 pKa = 11.84KK202 pKa = 9.37LNWRR206 pKa = 11.84NSLSGPDD213 pKa = 3.24AVNRR217 pKa = 11.84KK218 pKa = 8.62MSDD221 pKa = 3.24LVTKK225 pKa = 10.56AISSDD230 pKa = 3.46QYY232 pKa = 11.41LLSIDD237 pKa = 4.27FSAYY241 pKa = 9.55DD242 pKa = 3.59ASVKK246 pKa = 8.23TTLQKK251 pKa = 11.27ASFDD255 pKa = 4.02YY256 pKa = 10.65FKK258 pKa = 11.16SLYY261 pKa = 8.7QADD264 pKa = 3.97YY265 pKa = 10.94HH266 pKa = 7.17EE267 pKa = 5.85EE268 pKa = 3.74IDD270 pKa = 5.72DD271 pKa = 3.44IATRR275 pKa = 11.84FNTIGLVTPDD285 pKa = 4.54GIIDD289 pKa = 4.15GPHH292 pKa = 6.13GVPSGSTFTNEE303 pKa = 3.6VDD305 pKa = 4.53SVAQYY310 pKa = 10.99LIATEE315 pKa = 4.44YY316 pKa = 11.15GLSSDD321 pKa = 4.16NFDD324 pKa = 3.36IQGDD328 pKa = 3.66DD329 pKa = 3.24GAYY332 pKa = 10.01VVSDD336 pKa = 3.85PDD338 pKa = 3.57SLKK341 pKa = 11.1DD342 pKa = 3.46YY343 pKa = 9.98FRR345 pKa = 11.84KK346 pKa = 10.4YY347 pKa = 10.9NLVVNDD353 pKa = 4.01DD354 pKa = 3.4KK355 pKa = 11.94SYY357 pKa = 10.79ISKK360 pKa = 10.76DD361 pKa = 3.34YY362 pKa = 11.17LIYY365 pKa = 10.58LQNLYY370 pKa = 10.44HH371 pKa = 7.03KK372 pKa = 10.48DD373 pKa = 3.53YY374 pKa = 10.38MRR376 pKa = 11.84DD377 pKa = 3.66GIIGGIYY384 pKa = 6.86PTYY387 pKa = 9.93RR388 pKa = 11.84ALNRR392 pKa = 11.84IVYY395 pKa = 9.64LEE397 pKa = 4.28RR398 pKa = 11.84FTDD401 pKa = 4.15FLEE404 pKa = 5.65DD405 pKa = 3.77DD406 pKa = 5.03LKK408 pKa = 11.17GQDD411 pKa = 3.69YY412 pKa = 10.71FSLRR416 pKa = 11.84TISILEE422 pKa = 3.85NCKK425 pKa = 9.72FHH427 pKa = 7.3PLFEE431 pKa = 4.33EE432 pKa = 4.05LVKK435 pKa = 10.97YY436 pKa = 9.72VASLDD441 pKa = 3.76KK442 pKa = 11.49YY443 pKa = 9.59KK444 pKa = 10.98LKK446 pKa = 10.88YY447 pKa = 10.24SNSGLAKK454 pKa = 7.81YY455 pKa = 6.93TQRR458 pKa = 11.84ASQSSGVAGIFKK470 pKa = 10.25YY471 pKa = 10.3RR472 pKa = 11.84YY473 pKa = 8.78EE474 pKa = 5.18DD475 pKa = 4.19DD476 pKa = 4.15PKK478 pKa = 11.42GLNNFDD484 pKa = 3.69TVKK487 pKa = 10.64LLKK490 pKa = 10.39EE491 pKa = 4.01LL492 pKa = 3.98
MM1 pKa = 7.54NFQFLDD7 pKa = 3.6TLSVSKK13 pKa = 10.57QVEE16 pKa = 4.11NSVSSNLRR24 pKa = 11.84RR25 pKa = 11.84VVSGSEE31 pKa = 4.12DD32 pKa = 3.6VLTSPIVKK40 pKa = 9.98RR41 pKa = 11.84KK42 pKa = 9.96APEE45 pKa = 5.16DD46 pKa = 3.39ILKK49 pKa = 10.54GWDD52 pKa = 3.53EE53 pKa = 3.93IVKK56 pKa = 10.44EE57 pKa = 4.26NLSAMNADD65 pKa = 4.97LINLEE70 pKa = 4.49EE71 pKa = 4.34SNKK74 pKa = 10.52LKK76 pKa = 10.68FGPRR80 pKa = 11.84SIAAPWGEE88 pKa = 3.97RR89 pKa = 11.84TDD91 pKa = 4.21GVLSYY96 pKa = 10.31FAEE99 pKa = 4.58DD100 pKa = 3.68TLFTPPTVSKK110 pKa = 10.68VPRR113 pKa = 11.84RR114 pKa = 11.84NLRR117 pKa = 11.84PISEE121 pKa = 4.05AKK123 pKa = 10.7ALGFLKK129 pKa = 10.79NSTNSGLPYY138 pKa = 8.56YY139 pKa = 8.87TKK141 pKa = 10.25KK142 pKa = 11.04GKK144 pKa = 10.37VKK146 pKa = 10.28EE147 pKa = 4.5IYY149 pKa = 9.31STMQSPLLSRR159 pKa = 11.84KK160 pKa = 9.55DD161 pKa = 3.36PCVMFTRR168 pKa = 11.84TQEE171 pKa = 3.75QRR173 pKa = 11.84KK174 pKa = 6.66TRR176 pKa = 11.84TVWGYY181 pKa = 10.32PMADD185 pKa = 3.69TLRR188 pKa = 11.84EE189 pKa = 3.85MQYY192 pKa = 10.54YY193 pKa = 9.88EE194 pKa = 5.39PILGYY199 pKa = 7.76QRR201 pKa = 11.84KK202 pKa = 9.37LNWRR206 pKa = 11.84NSLSGPDD213 pKa = 3.24AVNRR217 pKa = 11.84KK218 pKa = 8.62MSDD221 pKa = 3.24LVTKK225 pKa = 10.56AISSDD230 pKa = 3.46QYY232 pKa = 11.41LLSIDD237 pKa = 4.27FSAYY241 pKa = 9.55DD242 pKa = 3.59ASVKK246 pKa = 8.23TTLQKK251 pKa = 11.27ASFDD255 pKa = 4.02YY256 pKa = 10.65FKK258 pKa = 11.16SLYY261 pKa = 8.7QADD264 pKa = 3.97YY265 pKa = 10.94HH266 pKa = 7.17EE267 pKa = 5.85EE268 pKa = 3.74IDD270 pKa = 5.72DD271 pKa = 3.44IATRR275 pKa = 11.84FNTIGLVTPDD285 pKa = 4.54GIIDD289 pKa = 4.15GPHH292 pKa = 6.13GVPSGSTFTNEE303 pKa = 3.6VDD305 pKa = 4.53SVAQYY310 pKa = 10.99LIATEE315 pKa = 4.44YY316 pKa = 11.15GLSSDD321 pKa = 4.16NFDD324 pKa = 3.36IQGDD328 pKa = 3.66DD329 pKa = 3.24GAYY332 pKa = 10.01VVSDD336 pKa = 3.85PDD338 pKa = 3.57SLKK341 pKa = 11.1DD342 pKa = 3.46YY343 pKa = 9.98FRR345 pKa = 11.84KK346 pKa = 10.4YY347 pKa = 10.9NLVVNDD353 pKa = 4.01DD354 pKa = 3.4KK355 pKa = 11.94SYY357 pKa = 10.79ISKK360 pKa = 10.76DD361 pKa = 3.34YY362 pKa = 11.17LIYY365 pKa = 10.58LQNLYY370 pKa = 10.44HH371 pKa = 7.03KK372 pKa = 10.48DD373 pKa = 3.53YY374 pKa = 10.38MRR376 pKa = 11.84DD377 pKa = 3.66GIIGGIYY384 pKa = 6.86PTYY387 pKa = 9.93RR388 pKa = 11.84ALNRR392 pKa = 11.84IVYY395 pKa = 9.64LEE397 pKa = 4.28RR398 pKa = 11.84FTDD401 pKa = 4.15FLEE404 pKa = 5.65DD405 pKa = 3.77DD406 pKa = 5.03LKK408 pKa = 11.17GQDD411 pKa = 3.69YY412 pKa = 10.71FSLRR416 pKa = 11.84TISILEE422 pKa = 3.85NCKK425 pKa = 9.72FHH427 pKa = 7.3PLFEE431 pKa = 4.33EE432 pKa = 4.05LVKK435 pKa = 10.97YY436 pKa = 9.72VASLDD441 pKa = 3.76KK442 pKa = 11.49YY443 pKa = 9.59KK444 pKa = 10.98LKK446 pKa = 10.88YY447 pKa = 10.24SNSGLAKK454 pKa = 7.81YY455 pKa = 6.93TQRR458 pKa = 11.84ASQSSGVAGIFKK470 pKa = 10.25YY471 pKa = 10.3RR472 pKa = 11.84YY473 pKa = 8.78EE474 pKa = 5.18DD475 pKa = 4.19DD476 pKa = 4.15PKK478 pKa = 11.42GLNNFDD484 pKa = 3.69TVKK487 pKa = 10.64LLKK490 pKa = 10.39EE491 pKa = 4.01LL492 pKa = 3.98
Molecular weight: 56.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLD7|A0A1L3KLD7_9VIRU Uncharacterized protein OS=Beihai picobirna-like virus 7 OX=1922524 PE=4 SV=1
MM1 pKa = 7.71IYY3 pKa = 10.27SKK5 pKa = 10.81SINKK9 pKa = 9.13VSLTKK14 pKa = 10.61LAAKK18 pKa = 10.16RR19 pKa = 11.84FQDD22 pKa = 3.62DD23 pKa = 4.49PYY25 pKa = 10.99VGMVLRR31 pKa = 11.84QSPEE35 pKa = 3.55FDD37 pKa = 3.26SFDD40 pKa = 3.47VNLRR44 pKa = 11.84SNPSNSDD51 pKa = 3.0SPRR54 pKa = 11.84NMSRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 8.08TSSNKK65 pKa = 9.37KK66 pKa = 8.36PKK68 pKa = 9.71KK69 pKa = 9.72KK70 pKa = 10.58KK71 pKa = 8.03PVQYY75 pKa = 10.27NGPKK79 pKa = 9.26AQRR82 pKa = 11.84ARR84 pKa = 11.84AQAQASSLAQGPFNLPPAAFVPGAPGFRR112 pKa = 11.84PATQPLLPVPTKK124 pKa = 10.85GVGALDD130 pKa = 4.33FAQSVVDD137 pKa = 4.66GIGRR141 pKa = 11.84GAAIVKK147 pKa = 9.12TGADD151 pKa = 3.85AIKK154 pKa = 10.67AIDD157 pKa = 4.07DD158 pKa = 3.62ATGGKK163 pKa = 9.13LRR165 pKa = 11.84PRR167 pKa = 11.84RR168 pKa = 11.84PRR170 pKa = 11.84NPDD173 pKa = 2.86RR174 pKa = 11.84VGGSGGEE181 pKa = 4.05SDD183 pKa = 3.89GSGGGGGGSKK193 pKa = 8.65STNLNNPNGGYY204 pKa = 9.73SYY206 pKa = 11.33TMMCNKK212 pKa = 9.57PVSQKK217 pKa = 10.18IEE219 pKa = 3.49ISTGVRR225 pKa = 11.84SGLIVNPGEE234 pKa = 4.25PNNDD238 pKa = 2.97SDD240 pKa = 4.81FSSLYY245 pKa = 10.35IMSGNLLKK253 pKa = 10.33KK254 pKa = 10.71DD255 pKa = 3.63SVDD258 pKa = 3.05TSFRR262 pKa = 11.84RR263 pKa = 11.84HH264 pKa = 6.0IEE266 pKa = 3.86TTIWPMVNQAVQYY279 pKa = 9.91NLNFEE284 pKa = 4.89YY285 pKa = 10.38ILSLTDD291 pKa = 3.48FNNWFYY297 pKa = 11.83DD298 pKa = 3.26MTKK301 pKa = 10.68ALEE304 pKa = 4.04VYY306 pKa = 9.78YY307 pKa = 10.96ALDD310 pKa = 3.32SVITYY315 pKa = 9.59GEE317 pKa = 4.04KK318 pKa = 10.58VEE320 pKa = 4.16NNNPGSRR327 pKa = 11.84YY328 pKa = 9.52LRR330 pKa = 11.84EE331 pKa = 4.74KK332 pKa = 8.68ITGPIRR338 pKa = 11.84LKK340 pKa = 10.9FNEE343 pKa = 4.5LRR345 pKa = 11.84NILNNQAIPPRR356 pKa = 11.84LIQLVRR362 pKa = 11.84YY363 pKa = 7.79MYY365 pKa = 10.18QYY367 pKa = 11.43YY368 pKa = 9.95KK369 pKa = 10.69FSEE372 pKa = 4.52LPGSPLYY379 pKa = 10.37RR380 pKa = 11.84LSPGFIFCNDD390 pKa = 3.1ANVGDD395 pKa = 4.56DD396 pKa = 4.36AYY398 pKa = 10.31IHH400 pKa = 6.53ALNTAMLDD408 pKa = 3.59KK409 pKa = 11.07LITNLTSHH417 pKa = 6.78SDD419 pKa = 3.47VHH421 pKa = 6.76NKK423 pKa = 9.33IYY425 pKa = 10.76KK426 pKa = 10.28AMPEE430 pKa = 3.64WHH432 pKa = 6.77VSGKK436 pKa = 9.77CLPPSCDD443 pKa = 2.77TAYY446 pKa = 10.76YY447 pKa = 10.28DD448 pKa = 3.77PSFRR452 pKa = 11.84TFWFNSCTSYY462 pKa = 11.03QVGNVVKK469 pKa = 8.31YY470 pKa = 7.94TRR472 pKa = 11.84KK473 pKa = 10.35GDD475 pKa = 3.24GSSNFRR481 pKa = 11.84YY482 pKa = 9.01WLYY485 pKa = 11.0TNNFDD490 pKa = 5.21GLFFALGSFNKK501 pKa = 8.12TTEE504 pKa = 3.76EE505 pKa = 4.26GTRR508 pKa = 11.84PGLWTPPSDD517 pKa = 3.41FSGIVGEE524 pKa = 4.38TNKK527 pKa = 10.22TSVQCFHH534 pKa = 7.31ADD536 pKa = 3.06KK537 pKa = 10.14EE538 pKa = 4.13IRR540 pKa = 11.84RR541 pKa = 11.84IVQLDD546 pKa = 3.46GTYY549 pKa = 10.97GKK551 pKa = 10.45ALQSDD556 pKa = 4.91NILVPYY562 pKa = 10.08QNLGGSGITIQLCHH576 pKa = 6.26NQDD579 pKa = 2.88GSTQVAQEE587 pKa = 4.43CTFNTYY593 pKa = 9.26KK594 pKa = 10.1QAVYY598 pKa = 10.24EE599 pKa = 4.12VTNWIFSII607 pKa = 4.36
MM1 pKa = 7.71IYY3 pKa = 10.27SKK5 pKa = 10.81SINKK9 pKa = 9.13VSLTKK14 pKa = 10.61LAAKK18 pKa = 10.16RR19 pKa = 11.84FQDD22 pKa = 3.62DD23 pKa = 4.49PYY25 pKa = 10.99VGMVLRR31 pKa = 11.84QSPEE35 pKa = 3.55FDD37 pKa = 3.26SFDD40 pKa = 3.47VNLRR44 pKa = 11.84SNPSNSDD51 pKa = 3.0SPRR54 pKa = 11.84NMSRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 8.08TSSNKK65 pKa = 9.37KK66 pKa = 8.36PKK68 pKa = 9.71KK69 pKa = 9.72KK70 pKa = 10.58KK71 pKa = 8.03PVQYY75 pKa = 10.27NGPKK79 pKa = 9.26AQRR82 pKa = 11.84ARR84 pKa = 11.84AQAQASSLAQGPFNLPPAAFVPGAPGFRR112 pKa = 11.84PATQPLLPVPTKK124 pKa = 10.85GVGALDD130 pKa = 4.33FAQSVVDD137 pKa = 4.66GIGRR141 pKa = 11.84GAAIVKK147 pKa = 9.12TGADD151 pKa = 3.85AIKK154 pKa = 10.67AIDD157 pKa = 4.07DD158 pKa = 3.62ATGGKK163 pKa = 9.13LRR165 pKa = 11.84PRR167 pKa = 11.84RR168 pKa = 11.84PRR170 pKa = 11.84NPDD173 pKa = 2.86RR174 pKa = 11.84VGGSGGEE181 pKa = 4.05SDD183 pKa = 3.89GSGGGGGGSKK193 pKa = 8.65STNLNNPNGGYY204 pKa = 9.73SYY206 pKa = 11.33TMMCNKK212 pKa = 9.57PVSQKK217 pKa = 10.18IEE219 pKa = 3.49ISTGVRR225 pKa = 11.84SGLIVNPGEE234 pKa = 4.25PNNDD238 pKa = 2.97SDD240 pKa = 4.81FSSLYY245 pKa = 10.35IMSGNLLKK253 pKa = 10.33KK254 pKa = 10.71DD255 pKa = 3.63SVDD258 pKa = 3.05TSFRR262 pKa = 11.84RR263 pKa = 11.84HH264 pKa = 6.0IEE266 pKa = 3.86TTIWPMVNQAVQYY279 pKa = 9.91NLNFEE284 pKa = 4.89YY285 pKa = 10.38ILSLTDD291 pKa = 3.48FNNWFYY297 pKa = 11.83DD298 pKa = 3.26MTKK301 pKa = 10.68ALEE304 pKa = 4.04VYY306 pKa = 9.78YY307 pKa = 10.96ALDD310 pKa = 3.32SVITYY315 pKa = 9.59GEE317 pKa = 4.04KK318 pKa = 10.58VEE320 pKa = 4.16NNNPGSRR327 pKa = 11.84YY328 pKa = 9.52LRR330 pKa = 11.84EE331 pKa = 4.74KK332 pKa = 8.68ITGPIRR338 pKa = 11.84LKK340 pKa = 10.9FNEE343 pKa = 4.5LRR345 pKa = 11.84NILNNQAIPPRR356 pKa = 11.84LIQLVRR362 pKa = 11.84YY363 pKa = 7.79MYY365 pKa = 10.18QYY367 pKa = 11.43YY368 pKa = 9.95KK369 pKa = 10.69FSEE372 pKa = 4.52LPGSPLYY379 pKa = 10.37RR380 pKa = 11.84LSPGFIFCNDD390 pKa = 3.1ANVGDD395 pKa = 4.56DD396 pKa = 4.36AYY398 pKa = 10.31IHH400 pKa = 6.53ALNTAMLDD408 pKa = 3.59KK409 pKa = 11.07LITNLTSHH417 pKa = 6.78SDD419 pKa = 3.47VHH421 pKa = 6.76NKK423 pKa = 9.33IYY425 pKa = 10.76KK426 pKa = 10.28AMPEE430 pKa = 3.64WHH432 pKa = 6.77VSGKK436 pKa = 9.77CLPPSCDD443 pKa = 2.77TAYY446 pKa = 10.76YY447 pKa = 10.28DD448 pKa = 3.77PSFRR452 pKa = 11.84TFWFNSCTSYY462 pKa = 11.03QVGNVVKK469 pKa = 8.31YY470 pKa = 7.94TRR472 pKa = 11.84KK473 pKa = 10.35GDD475 pKa = 3.24GSSNFRR481 pKa = 11.84YY482 pKa = 9.01WLYY485 pKa = 11.0TNNFDD490 pKa = 5.21GLFFALGSFNKK501 pKa = 8.12TTEE504 pKa = 3.76EE505 pKa = 4.26GTRR508 pKa = 11.84PGLWTPPSDD517 pKa = 3.41FSGIVGEE524 pKa = 4.38TNKK527 pKa = 10.22TSVQCFHH534 pKa = 7.31ADD536 pKa = 3.06KK537 pKa = 10.14EE538 pKa = 4.13IRR540 pKa = 11.84RR541 pKa = 11.84IVQLDD546 pKa = 3.46GTYY549 pKa = 10.97GKK551 pKa = 10.45ALQSDD556 pKa = 4.91NILVPYY562 pKa = 10.08QNLGGSGITIQLCHH576 pKa = 6.26NQDD579 pKa = 2.88GSTQVAQEE587 pKa = 4.43CTFNTYY593 pKa = 9.26KK594 pKa = 10.1QAVYY598 pKa = 10.24EE599 pKa = 4.12VTNWIFSII607 pKa = 4.36
Molecular weight: 67.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1099 |
492 |
607 |
549.5 |
61.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.369 ± 0.322 | 0.91 ± 0.33 |
6.733 ± 1.05 | 4.004 ± 0.707 |
4.459 ± 0.125 | 7.188 ± 1.116 |
1.001 ± 0.123 | 5.187 ± 0.064 |
6.824 ± 0.59 | 8.371 ± 1.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.729 ± 0.067 | 6.46 ± 1.038 |
5.46 ± 0.781 | 3.731 ± 0.314 |
5.096 ± 0.009 | 9.008 ± 0.224 |
5.823 ± 0.087 | 5.914 ± 0.12 |
1.001 ± 0.123 | 5.732 ± 0.64 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
