
Candidatus Izimaplasma sp. HR1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Tenericutes incertae sedis; Candidatus Izimaplasma; unclassified Candidatus Izimaplasma
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
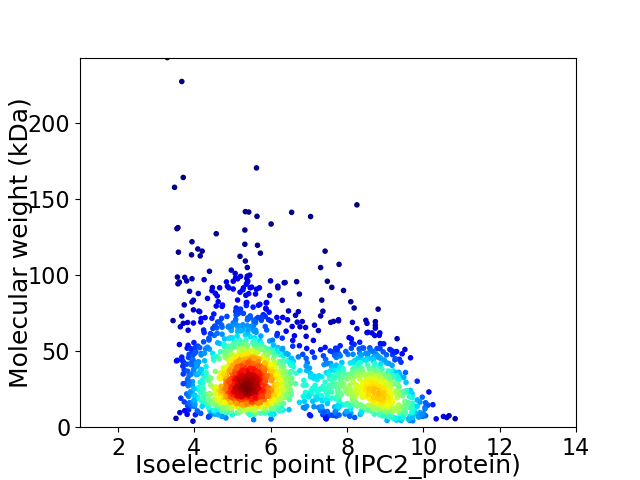
Virtual 2D-PAGE plot for 1811 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A088T012|A0A088T012_9BACT Recombination protein RecR OS=Candidatus Izimaplasma sp. HR1 OX=1541959 GN=recR PE=3 SV=1
MM1 pKa = 7.39KK2 pKa = 10.37KK3 pKa = 10.3VLTLLLVFTLGLTLASCTDD22 pKa = 3.29QEE24 pKa = 4.46AATNVADD31 pKa = 4.22AAQKK35 pKa = 10.86LSLEE39 pKa = 4.37VSDD42 pKa = 4.22PTAVTSDD49 pKa = 2.62ITLPTEE55 pKa = 4.35GLHH58 pKa = 6.47EE59 pKa = 4.38STISWEE65 pKa = 4.14SDD67 pKa = 2.69NAAISAAGVVTRR79 pKa = 11.84PAIGSDD85 pKa = 3.36DD86 pKa = 3.56VTVILTATFTIGKK99 pKa = 7.05EE100 pKa = 4.11TTTKK104 pKa = 10.56EE105 pKa = 3.83FTFKK109 pKa = 10.73VLAAVPSVAVTAAVFEE125 pKa = 4.61SSATAVGDD133 pKa = 3.88VVEE136 pKa = 4.26LTGIVVGLVKK146 pKa = 10.11TYY148 pKa = 10.44GYY150 pKa = 10.3EE151 pKa = 3.91IYY153 pKa = 10.47DD154 pKa = 3.66GTGFTYY160 pKa = 10.75VNEE163 pKa = 4.38FQDD166 pKa = 3.9PTVSIGDD173 pKa = 3.83EE174 pKa = 4.14VTVMGEE180 pKa = 3.53KK181 pKa = 10.38GIYY184 pKa = 9.86NNGSEE189 pKa = 4.2LVEE192 pKa = 4.1IQSTVVNSTGNALPNFTEE210 pKa = 4.26TTIGEE215 pKa = 4.37IFAQDD220 pKa = 3.88PDD222 pKa = 3.94TTDD225 pKa = 2.65WYY227 pKa = 11.02HH228 pKa = 8.59DD229 pKa = 3.54MFNAEE234 pKa = 4.06GVIVVQGDD242 pKa = 3.43YY243 pKa = 11.62DD244 pKa = 3.58NAYY247 pKa = 10.65LSWFDD252 pKa = 3.03QDD254 pKa = 4.0LNLRR258 pKa = 11.84QMEE261 pKa = 4.74IYY263 pKa = 9.97YY264 pKa = 10.48KK265 pKa = 10.68SYY267 pKa = 11.63DD268 pKa = 3.78DD269 pKa = 4.58AKK271 pKa = 10.67LAAVKK276 pKa = 10.34ALEE279 pKa = 4.3GKK281 pKa = 10.03KK282 pKa = 10.11VEE284 pKa = 4.8LDD286 pKa = 4.6FILKK290 pKa = 10.05DD291 pKa = 3.37YY292 pKa = 10.92SYY294 pKa = 11.79GSFRR298 pKa = 11.84LTVNSTGEE306 pKa = 4.07VNEE309 pKa = 3.82ATIYY313 pKa = 10.53TDD315 pKa = 3.68AEE317 pKa = 4.15RR318 pKa = 11.84AKK320 pKa = 10.89LAVAYY325 pKa = 10.54NDD327 pKa = 4.3LFFNSLGDD335 pKa = 3.76EE336 pKa = 4.55VVASLDD342 pKa = 3.98LPTDD346 pKa = 3.35SDD348 pKa = 4.5FVTKK352 pKa = 10.86YY353 pKa = 11.29DD354 pKa = 3.76DD355 pKa = 3.65TEE357 pKa = 4.08EE358 pKa = 5.16ANMPIEE364 pKa = 4.53LVWTSSNEE372 pKa = 3.72VVIANDD378 pKa = 3.72GTVTLPVGATADD390 pKa = 3.57VEE392 pKa = 4.53LTVTATVGDD401 pKa = 3.99VSAHH405 pKa = 4.42KK406 pKa = 9.38TYY408 pKa = 10.39TVTVADD414 pKa = 4.78ADD416 pKa = 4.11TLTTNTVAEE425 pKa = 4.78VIAMEE430 pKa = 4.88DD431 pKa = 3.84GEE433 pKa = 4.83TVIVEE438 pKa = 4.68AIVSGLRR445 pKa = 11.84YY446 pKa = 10.14GNPFVQDD453 pKa = 3.29ADD455 pKa = 3.77GTAMFIYY462 pKa = 9.82TKK464 pKa = 10.74LDD466 pKa = 3.33VKK468 pKa = 11.13VGDD471 pKa = 4.06KK472 pKa = 10.85VWIQGEE478 pKa = 4.17LATFSSYY485 pKa = 11.4GNALKK490 pKa = 10.58QLTRR494 pKa = 11.84GVLVKK499 pKa = 9.83TVSTGNALTYY509 pKa = 9.1NTTTTAAEE517 pKa = 3.66LAAAINTMHH526 pKa = 6.99SDD528 pKa = 3.5SFTMTLKK535 pKa = 10.64FLEE538 pKa = 4.5EE539 pKa = 3.87DD540 pKa = 2.95FGYY543 pKa = 10.95YY544 pKa = 10.03FFEE547 pKa = 6.18AYY549 pKa = 10.49VDD551 pKa = 5.29GDD553 pKa = 3.86DD554 pKa = 4.12ADD556 pKa = 3.87NNVRR560 pKa = 11.84IKK562 pKa = 10.27MDD564 pKa = 3.86TDD566 pKa = 3.71WFPTMEE572 pKa = 4.35TYY574 pKa = 11.21LDD576 pKa = 4.02GSDD579 pKa = 3.62MIEE582 pKa = 3.83ISFVLYY588 pKa = 10.27DD589 pKa = 3.13VHH591 pKa = 7.96YY592 pKa = 10.2DD593 pKa = 3.39EE594 pKa = 6.02ARR596 pKa = 11.84IVPVAFPGLTNADD609 pKa = 3.64RR610 pKa = 11.84LDD612 pKa = 3.49IVKK615 pKa = 10.42GMIALEE621 pKa = 4.01EE622 pKa = 4.52FIVLDD627 pKa = 4.07VNLLTYY633 pKa = 10.86LPTFDD638 pKa = 5.98AAITWVSANTAMDD651 pKa = 3.55AEE653 pKa = 4.9GNVTRR658 pKa = 11.84PVAGEE663 pKa = 4.16DD664 pKa = 3.52NVEE667 pKa = 4.13GDD669 pKa = 3.77LTASIVIGSDD679 pKa = 2.97AAVDD683 pKa = 3.59EE684 pKa = 5.23VYY686 pKa = 10.3TVTVVASPTDD696 pKa = 3.18AMTVAEE702 pKa = 5.1VYY704 pKa = 10.35AAGYY708 pKa = 9.92SDD710 pKa = 4.44YY711 pKa = 11.13FGDD714 pKa = 3.8YY715 pKa = 7.83THH717 pKa = 6.91VYY719 pKa = 7.46VTGVVAGLTDD729 pKa = 4.21DD730 pKa = 5.05GYY732 pKa = 11.62VIQDD736 pKa = 3.57TVSGKK741 pKa = 10.3FLVVEE746 pKa = 4.47DD747 pKa = 4.59FDD749 pKa = 5.12NEE751 pKa = 4.37VEE753 pKa = 4.63VGQEE757 pKa = 3.69LNLEE761 pKa = 4.19GFYY764 pKa = 10.6NASYY768 pKa = 10.66EE769 pKa = 4.36VIKK772 pKa = 11.02LRR774 pKa = 11.84DD775 pKa = 3.29VFVFMFVSEE784 pKa = 4.38GNALNLEE791 pKa = 4.25LTDD794 pKa = 4.01AVAMDD799 pKa = 4.09WATFDD804 pKa = 4.07RR805 pKa = 11.84NDD807 pKa = 3.5YY808 pKa = 8.76QTGEE812 pKa = 4.73LIAITAPWATLYY824 pKa = 11.19SGDD827 pKa = 3.42TSYY830 pKa = 11.64LRR832 pKa = 11.84LGNGDD837 pKa = 3.47GLEE840 pKa = 4.14NAKK843 pKa = 10.72SFDD846 pKa = 3.32GKK848 pKa = 11.08YY849 pKa = 10.56VGLQNGANEE858 pKa = 4.33LNLTGTLADD867 pKa = 4.24IFDD870 pKa = 4.52GGQTSTEE877 pKa = 4.14YY878 pKa = 11.39ADD880 pKa = 3.21VTIYY884 pKa = 11.15VLIYY888 pKa = 9.23DD889 pKa = 4.1TTSSYY894 pKa = 11.85NKK896 pKa = 10.15FIILSDD902 pKa = 3.68DD903 pKa = 4.36FIVVTPP909 pKa = 4.54
MM1 pKa = 7.39KK2 pKa = 10.37KK3 pKa = 10.3VLTLLLVFTLGLTLASCTDD22 pKa = 3.29QEE24 pKa = 4.46AATNVADD31 pKa = 4.22AAQKK35 pKa = 10.86LSLEE39 pKa = 4.37VSDD42 pKa = 4.22PTAVTSDD49 pKa = 2.62ITLPTEE55 pKa = 4.35GLHH58 pKa = 6.47EE59 pKa = 4.38STISWEE65 pKa = 4.14SDD67 pKa = 2.69NAAISAAGVVTRR79 pKa = 11.84PAIGSDD85 pKa = 3.36DD86 pKa = 3.56VTVILTATFTIGKK99 pKa = 7.05EE100 pKa = 4.11TTTKK104 pKa = 10.56EE105 pKa = 3.83FTFKK109 pKa = 10.73VLAAVPSVAVTAAVFEE125 pKa = 4.61SSATAVGDD133 pKa = 3.88VVEE136 pKa = 4.26LTGIVVGLVKK146 pKa = 10.11TYY148 pKa = 10.44GYY150 pKa = 10.3EE151 pKa = 3.91IYY153 pKa = 10.47DD154 pKa = 3.66GTGFTYY160 pKa = 10.75VNEE163 pKa = 4.38FQDD166 pKa = 3.9PTVSIGDD173 pKa = 3.83EE174 pKa = 4.14VTVMGEE180 pKa = 3.53KK181 pKa = 10.38GIYY184 pKa = 9.86NNGSEE189 pKa = 4.2LVEE192 pKa = 4.1IQSTVVNSTGNALPNFTEE210 pKa = 4.26TTIGEE215 pKa = 4.37IFAQDD220 pKa = 3.88PDD222 pKa = 3.94TTDD225 pKa = 2.65WYY227 pKa = 11.02HH228 pKa = 8.59DD229 pKa = 3.54MFNAEE234 pKa = 4.06GVIVVQGDD242 pKa = 3.43YY243 pKa = 11.62DD244 pKa = 3.58NAYY247 pKa = 10.65LSWFDD252 pKa = 3.03QDD254 pKa = 4.0LNLRR258 pKa = 11.84QMEE261 pKa = 4.74IYY263 pKa = 9.97YY264 pKa = 10.48KK265 pKa = 10.68SYY267 pKa = 11.63DD268 pKa = 3.78DD269 pKa = 4.58AKK271 pKa = 10.67LAAVKK276 pKa = 10.34ALEE279 pKa = 4.3GKK281 pKa = 10.03KK282 pKa = 10.11VEE284 pKa = 4.8LDD286 pKa = 4.6FILKK290 pKa = 10.05DD291 pKa = 3.37YY292 pKa = 10.92SYY294 pKa = 11.79GSFRR298 pKa = 11.84LTVNSTGEE306 pKa = 4.07VNEE309 pKa = 3.82ATIYY313 pKa = 10.53TDD315 pKa = 3.68AEE317 pKa = 4.15RR318 pKa = 11.84AKK320 pKa = 10.89LAVAYY325 pKa = 10.54NDD327 pKa = 4.3LFFNSLGDD335 pKa = 3.76EE336 pKa = 4.55VVASLDD342 pKa = 3.98LPTDD346 pKa = 3.35SDD348 pKa = 4.5FVTKK352 pKa = 10.86YY353 pKa = 11.29DD354 pKa = 3.76DD355 pKa = 3.65TEE357 pKa = 4.08EE358 pKa = 5.16ANMPIEE364 pKa = 4.53LVWTSSNEE372 pKa = 3.72VVIANDD378 pKa = 3.72GTVTLPVGATADD390 pKa = 3.57VEE392 pKa = 4.53LTVTATVGDD401 pKa = 3.99VSAHH405 pKa = 4.42KK406 pKa = 9.38TYY408 pKa = 10.39TVTVADD414 pKa = 4.78ADD416 pKa = 4.11TLTTNTVAEE425 pKa = 4.78VIAMEE430 pKa = 4.88DD431 pKa = 3.84GEE433 pKa = 4.83TVIVEE438 pKa = 4.68AIVSGLRR445 pKa = 11.84YY446 pKa = 10.14GNPFVQDD453 pKa = 3.29ADD455 pKa = 3.77GTAMFIYY462 pKa = 9.82TKK464 pKa = 10.74LDD466 pKa = 3.33VKK468 pKa = 11.13VGDD471 pKa = 4.06KK472 pKa = 10.85VWIQGEE478 pKa = 4.17LATFSSYY485 pKa = 11.4GNALKK490 pKa = 10.58QLTRR494 pKa = 11.84GVLVKK499 pKa = 9.83TVSTGNALTYY509 pKa = 9.1NTTTTAAEE517 pKa = 3.66LAAAINTMHH526 pKa = 6.99SDD528 pKa = 3.5SFTMTLKK535 pKa = 10.64FLEE538 pKa = 4.5EE539 pKa = 3.87DD540 pKa = 2.95FGYY543 pKa = 10.95YY544 pKa = 10.03FFEE547 pKa = 6.18AYY549 pKa = 10.49VDD551 pKa = 5.29GDD553 pKa = 3.86DD554 pKa = 4.12ADD556 pKa = 3.87NNVRR560 pKa = 11.84IKK562 pKa = 10.27MDD564 pKa = 3.86TDD566 pKa = 3.71WFPTMEE572 pKa = 4.35TYY574 pKa = 11.21LDD576 pKa = 4.02GSDD579 pKa = 3.62MIEE582 pKa = 3.83ISFVLYY588 pKa = 10.27DD589 pKa = 3.13VHH591 pKa = 7.96YY592 pKa = 10.2DD593 pKa = 3.39EE594 pKa = 6.02ARR596 pKa = 11.84IVPVAFPGLTNADD609 pKa = 3.64RR610 pKa = 11.84LDD612 pKa = 3.49IVKK615 pKa = 10.42GMIALEE621 pKa = 4.01EE622 pKa = 4.52FIVLDD627 pKa = 4.07VNLLTYY633 pKa = 10.86LPTFDD638 pKa = 5.98AAITWVSANTAMDD651 pKa = 3.55AEE653 pKa = 4.9GNVTRR658 pKa = 11.84PVAGEE663 pKa = 4.16DD664 pKa = 3.52NVEE667 pKa = 4.13GDD669 pKa = 3.77LTASIVIGSDD679 pKa = 2.97AAVDD683 pKa = 3.59EE684 pKa = 5.23VYY686 pKa = 10.3TVTVVASPTDD696 pKa = 3.18AMTVAEE702 pKa = 5.1VYY704 pKa = 10.35AAGYY708 pKa = 9.92SDD710 pKa = 4.44YY711 pKa = 11.13FGDD714 pKa = 3.8YY715 pKa = 7.83THH717 pKa = 6.91VYY719 pKa = 7.46VTGVVAGLTDD729 pKa = 4.21DD730 pKa = 5.05GYY732 pKa = 11.62VIQDD736 pKa = 3.57TVSGKK741 pKa = 10.3FLVVEE746 pKa = 4.47DD747 pKa = 4.59FDD749 pKa = 5.12NEE751 pKa = 4.37VEE753 pKa = 4.63VGQEE757 pKa = 3.69LNLEE761 pKa = 4.19GFYY764 pKa = 10.6NASYY768 pKa = 10.66EE769 pKa = 4.36VIKK772 pKa = 11.02LRR774 pKa = 11.84DD775 pKa = 3.29VFVFMFVSEE784 pKa = 4.38GNALNLEE791 pKa = 4.25LTDD794 pKa = 4.01AVAMDD799 pKa = 4.09WATFDD804 pKa = 4.07RR805 pKa = 11.84NDD807 pKa = 3.5YY808 pKa = 8.76QTGEE812 pKa = 4.73LIAITAPWATLYY824 pKa = 11.19SGDD827 pKa = 3.42TSYY830 pKa = 11.64LRR832 pKa = 11.84LGNGDD837 pKa = 3.47GLEE840 pKa = 4.14NAKK843 pKa = 10.72SFDD846 pKa = 3.32GKK848 pKa = 11.08YY849 pKa = 10.56VGLQNGANEE858 pKa = 4.33LNLTGTLADD867 pKa = 4.24IFDD870 pKa = 4.52GGQTSTEE877 pKa = 4.14YY878 pKa = 11.39ADD880 pKa = 3.21VTIYY884 pKa = 11.15VLIYY888 pKa = 9.23DD889 pKa = 4.1TTSSYY894 pKa = 11.85NKK896 pKa = 10.15FIILSDD902 pKa = 3.68DD903 pKa = 4.36FIVVTPP909 pKa = 4.54
Molecular weight: 98.49 kDa
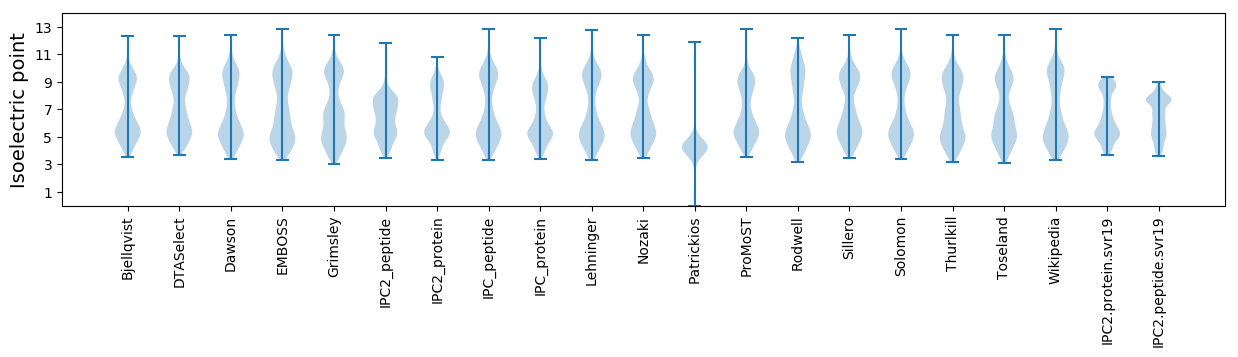
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A088SZL1|A0A088SZL1_9BACT Probable membrane transporter protein OS=Candidatus Izimaplasma sp. HR1 OX=1541959 GN=KQ51_00185 PE=3 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.41KK4 pKa = 10.39SMKK7 pKa = 9.28VKK9 pKa = 9.97QRR11 pKa = 11.84RR12 pKa = 11.84GSKK15 pKa = 10.15FSARR19 pKa = 11.84EE20 pKa = 3.52YY21 pKa = 9.96TRR23 pKa = 11.84CEE25 pKa = 3.78RR26 pKa = 11.84CGRR29 pKa = 11.84PHH31 pKa = 6.43AVYY34 pKa = 10.39RR35 pKa = 11.84KK36 pKa = 9.55FKK38 pKa = 10.41LCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.94LAYY49 pKa = 10.67LGQIPGVKK57 pKa = 9.32KK58 pKa = 10.9ASWW61 pKa = 3.03
MM1 pKa = 7.61AKK3 pKa = 10.41KK4 pKa = 10.39SMKK7 pKa = 9.28VKK9 pKa = 9.97QRR11 pKa = 11.84RR12 pKa = 11.84GSKK15 pKa = 10.15FSARR19 pKa = 11.84EE20 pKa = 3.52YY21 pKa = 9.96TRR23 pKa = 11.84CEE25 pKa = 3.78RR26 pKa = 11.84CGRR29 pKa = 11.84PHH31 pKa = 6.43AVYY34 pKa = 10.39RR35 pKa = 11.84KK36 pKa = 9.55FKK38 pKa = 10.41LCRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.94LAYY49 pKa = 10.67LGQIPGVKK57 pKa = 9.32KK58 pKa = 10.9ASWW61 pKa = 3.03
Molecular weight: 7.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
571813 |
34 |
2255 |
315.7 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.256 ± 0.06 | 0.761 ± 0.021 |
6.028 ± 0.052 | 7.213 ± 0.061 |
4.943 ± 0.054 | 6.155 ± 0.06 |
1.699 ± 0.022 | 9.378 ± 0.058 |
7.712 ± 0.088 | 9.614 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.513 ± 0.025 | 5.808 ± 0.055 |
2.817 ± 0.03 | 2.486 ± 0.026 |
3.569 ± 0.042 | 6.089 ± 0.043 |
5.658 ± 0.048 | 6.986 ± 0.045 |
0.658 ± 0.019 | 4.654 ± 0.052 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
