
Clostridium tetani (strain Massachusetts / E88)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; Clostridium tetani
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
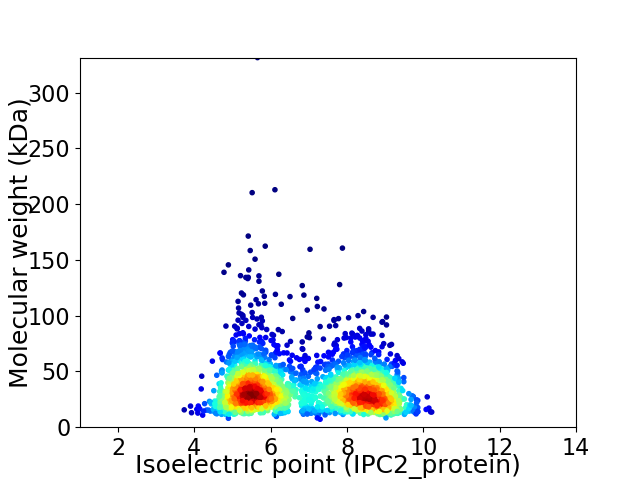
Virtual 2D-PAGE plot for 2415 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
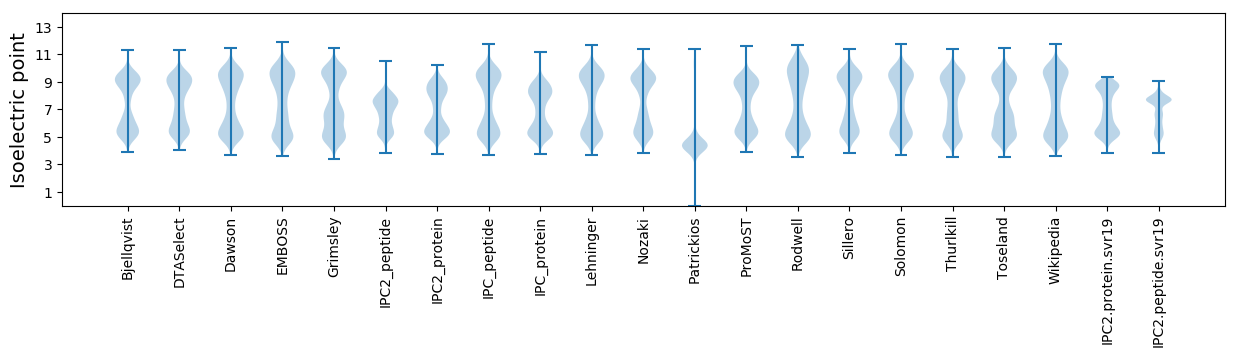
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q898L8|Q898L8_CLOTE Glycerophosphoryl diester phosphodiesterase OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=CTC_00427 PE=4 SV=1
MM1 pKa = 7.3FLCFYY6 pKa = 10.91VLTKK10 pKa = 10.75LEE12 pKa = 3.82ISNNIIYY19 pKa = 10.48EE20 pKa = 3.98SDD22 pKa = 3.24NQYY25 pKa = 10.88QLPGGIYY32 pKa = 8.42MKK34 pKa = 10.29NISVIYY40 pKa = 9.79WSGTGNTEE48 pKa = 4.04AMAEE52 pKa = 3.99ALAEE56 pKa = 4.19GAKK59 pKa = 10.43AAGGQVKK66 pKa = 9.85LVSVDD71 pKa = 3.64EE72 pKa = 4.28VSEE75 pKa = 4.78DD76 pKa = 3.63DD77 pKa = 4.43VKK79 pKa = 11.45NADD82 pKa = 4.12AVALGCPSMGDD93 pKa = 3.28EE94 pKa = 4.17VLEE97 pKa = 4.47EE98 pKa = 4.52GSMEE102 pKa = 4.03PFIEE106 pKa = 4.26QTSNLYY112 pKa = 9.98KK113 pKa = 10.5GKK115 pKa = 10.04NVALFGSYY123 pKa = 9.05GWGSGQWMEE132 pKa = 4.07DD133 pKa = 2.53WQARR137 pKa = 11.84MEE139 pKa = 4.62GYY141 pKa = 9.77GANLVEE147 pKa = 5.98DD148 pKa = 4.33GLIINYY154 pKa = 8.75TPDD157 pKa = 3.82DD158 pKa = 4.23EE159 pKa = 5.58GLEE162 pKa = 4.12KK163 pKa = 10.59CKK165 pKa = 10.4QLGEE169 pKa = 4.4SLVKK173 pKa = 10.76AA174 pKa = 4.39
MM1 pKa = 7.3FLCFYY6 pKa = 10.91VLTKK10 pKa = 10.75LEE12 pKa = 3.82ISNNIIYY19 pKa = 10.48EE20 pKa = 3.98SDD22 pKa = 3.24NQYY25 pKa = 10.88QLPGGIYY32 pKa = 8.42MKK34 pKa = 10.29NISVIYY40 pKa = 9.79WSGTGNTEE48 pKa = 4.04AMAEE52 pKa = 3.99ALAEE56 pKa = 4.19GAKK59 pKa = 10.43AAGGQVKK66 pKa = 9.85LVSVDD71 pKa = 3.64EE72 pKa = 4.28VSEE75 pKa = 4.78DD76 pKa = 3.63DD77 pKa = 4.43VKK79 pKa = 11.45NADD82 pKa = 4.12AVALGCPSMGDD93 pKa = 3.28EE94 pKa = 4.17VLEE97 pKa = 4.47EE98 pKa = 4.52GSMEE102 pKa = 4.03PFIEE106 pKa = 4.26QTSNLYY112 pKa = 9.98KK113 pKa = 10.5GKK115 pKa = 10.04NVALFGSYY123 pKa = 9.05GWGSGQWMEE132 pKa = 4.07DD133 pKa = 2.53WQARR137 pKa = 11.84MEE139 pKa = 4.62GYY141 pKa = 9.77GANLVEE147 pKa = 5.98DD148 pKa = 4.33GLIINYY154 pKa = 8.75TPDD157 pKa = 3.82DD158 pKa = 4.23EE159 pKa = 5.58GLEE162 pKa = 4.12KK163 pKa = 10.59CKK165 pKa = 10.4QLGEE169 pKa = 4.4SLVKK173 pKa = 10.76AA174 pKa = 4.39
Molecular weight: 18.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q898L3|HIS2_CLOTE Phosphoribosyl-ATP pyrophosphatase OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=hisE PE=3 SV=1
MM1 pKa = 7.41YY2 pKa = 9.67YY3 pKa = 10.45WNMNCGDD10 pKa = 3.87CFYY13 pKa = 10.47RR14 pKa = 11.84QQEE17 pKa = 3.96AMEE20 pKa = 4.49LNRR23 pKa = 11.84RR24 pKa = 11.84WRR26 pKa = 11.84PWRR29 pKa = 11.84VISGRR34 pKa = 11.84VTSGLGVSSWAPNRR48 pKa = 11.84LDD50 pKa = 3.49LFARR54 pKa = 11.84GSNRR58 pKa = 11.84QLIHH62 pKa = 6.53KK63 pKa = 8.87WRR65 pKa = 11.84EE66 pKa = 3.35NSRR69 pKa = 11.84WSNWEE74 pKa = 3.75DD75 pKa = 3.18LGGVLTSSPSAVSWGRR91 pKa = 11.84NRR93 pKa = 11.84IDD95 pKa = 3.27VVVRR99 pKa = 11.84GTDD102 pKa = 2.63NAMYY106 pKa = 9.75HH107 pKa = 5.75KK108 pKa = 9.23WWNGSRR114 pKa = 11.84WSDD117 pKa = 3.06WEE119 pKa = 4.15SLGGVLTSAPSICSWASNRR138 pKa = 11.84LDD140 pKa = 3.49CFVRR144 pKa = 11.84GTDD147 pKa = 3.08NQLYY151 pKa = 8.96HH152 pKa = 6.61KK153 pKa = 9.05WWDD156 pKa = 3.21GSRR159 pKa = 11.84WHH161 pKa = 7.1DD162 pKa = 3.37WEE164 pKa = 5.0ALGGNLTSGPGCVSWGPNRR183 pKa = 11.84IDD185 pKa = 3.08VFARR189 pKa = 11.84GRR191 pKa = 11.84NNTLIHH197 pKa = 6.02KK198 pKa = 8.67WWDD201 pKa = 3.17GSRR204 pKa = 11.84WSDD207 pKa = 3.26WEE209 pKa = 4.13DD210 pKa = 3.18LGGNLSSAPCASSRR224 pKa = 11.84GRR226 pKa = 11.84NRR228 pKa = 11.84IDD230 pKa = 2.97VFARR234 pKa = 11.84GRR236 pKa = 11.84NNQLIYY242 pKa = 10.95KK243 pKa = 8.46NWNGSRR249 pKa = 11.84WSNWQNLGGFLTSPPVSISPTPNRR273 pKa = 11.84IEE275 pKa = 3.97VYY277 pKa = 10.17ARR279 pKa = 11.84GRR281 pKa = 11.84NGIIINRR288 pKa = 11.84NFSGG292 pKa = 3.42
MM1 pKa = 7.41YY2 pKa = 9.67YY3 pKa = 10.45WNMNCGDD10 pKa = 3.87CFYY13 pKa = 10.47RR14 pKa = 11.84QQEE17 pKa = 3.96AMEE20 pKa = 4.49LNRR23 pKa = 11.84RR24 pKa = 11.84WRR26 pKa = 11.84PWRR29 pKa = 11.84VISGRR34 pKa = 11.84VTSGLGVSSWAPNRR48 pKa = 11.84LDD50 pKa = 3.49LFARR54 pKa = 11.84GSNRR58 pKa = 11.84QLIHH62 pKa = 6.53KK63 pKa = 8.87WRR65 pKa = 11.84EE66 pKa = 3.35NSRR69 pKa = 11.84WSNWEE74 pKa = 3.75DD75 pKa = 3.18LGGVLTSSPSAVSWGRR91 pKa = 11.84NRR93 pKa = 11.84IDD95 pKa = 3.27VVVRR99 pKa = 11.84GTDD102 pKa = 2.63NAMYY106 pKa = 9.75HH107 pKa = 5.75KK108 pKa = 9.23WWNGSRR114 pKa = 11.84WSDD117 pKa = 3.06WEE119 pKa = 4.15SLGGVLTSAPSICSWASNRR138 pKa = 11.84LDD140 pKa = 3.49CFVRR144 pKa = 11.84GTDD147 pKa = 3.08NQLYY151 pKa = 8.96HH152 pKa = 6.61KK153 pKa = 9.05WWDD156 pKa = 3.21GSRR159 pKa = 11.84WHH161 pKa = 7.1DD162 pKa = 3.37WEE164 pKa = 5.0ALGGNLTSGPGCVSWGPNRR183 pKa = 11.84IDD185 pKa = 3.08VFARR189 pKa = 11.84GRR191 pKa = 11.84NNTLIHH197 pKa = 6.02KK198 pKa = 8.67WWDD201 pKa = 3.17GSRR204 pKa = 11.84WSDD207 pKa = 3.26WEE209 pKa = 4.13DD210 pKa = 3.18LGGNLSSAPCASSRR224 pKa = 11.84GRR226 pKa = 11.84NRR228 pKa = 11.84IDD230 pKa = 2.97VFARR234 pKa = 11.84GRR236 pKa = 11.84NNQLIYY242 pKa = 10.95KK243 pKa = 8.46NWNGSRR249 pKa = 11.84WSNWQNLGGFLTSPPVSISPTPNRR273 pKa = 11.84IEE275 pKa = 3.97VYY277 pKa = 10.17ARR279 pKa = 11.84GRR281 pKa = 11.84NGIIINRR288 pKa = 11.84NFSGG292 pKa = 3.42
Molecular weight: 33.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
809352 |
60 |
2838 |
335.1 |
37.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.229 ± 0.058 | 1.127 ± 0.017 |
5.389 ± 0.037 | 7.644 ± 0.059 |
4.413 ± 0.04 | 6.317 ± 0.05 |
1.342 ± 0.019 | 10.456 ± 0.064 |
9.962 ± 0.059 | 9.006 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.606 ± 0.022 | 6.555 ± 0.052 |
2.746 ± 0.027 | 2.146 ± 0.019 |
3.285 ± 0.037 | 6.163 ± 0.033 |
4.673 ± 0.036 | 6.165 ± 0.042 |
0.642 ± 0.015 | 4.131 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
