
Candidatus Nitrosotalea sinensis
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Thaumarchaeota incertae sedis; Candidatus Nitrosotalea
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
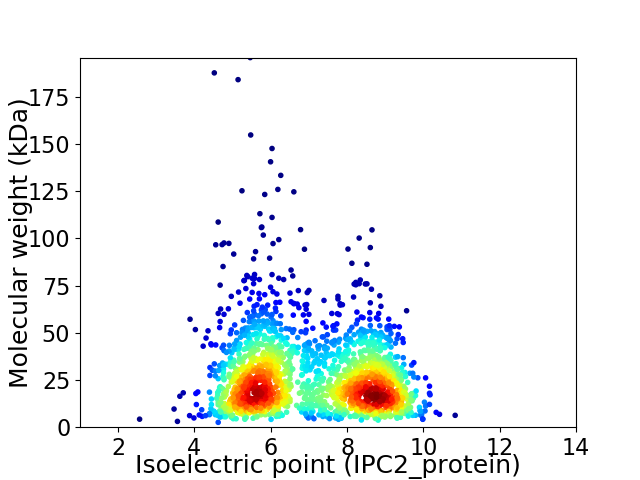
Virtual 2D-PAGE plot for 1881 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H1EHI7|A0A2H1EHI7_9ARCH Ribokinase-like domain-containing protein OS=Candidatus Nitrosotalea sinensis OX=1499975 GN=NSIN_30110 PE=4 SV=1
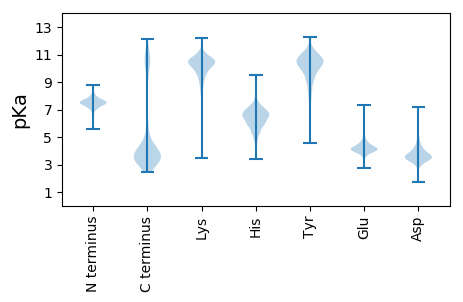
MM1 pKa = 7.33TNINAKK7 pKa = 8.84ATGPSGAPVTFSATATDD24 pKa = 4.14IVDD27 pKa = 3.5GTDD30 pKa = 4.4PVTCDD35 pKa = 3.29HH36 pKa = 6.84NSGDD40 pKa = 4.16TYY42 pKa = 11.19PLGSTTVSCSSTDD55 pKa = 3.29HH56 pKa = 7.08AGNTSHH62 pKa = 7.38ASFTITVAEE71 pKa = 4.44TTAPSITLPGTITQEE86 pKa = 3.69ATGPSGAPVTFSATATDD103 pKa = 4.14IVDD106 pKa = 3.5GTDD109 pKa = 3.49PVTCTPASGSTFALGSTTVNCSSTDD134 pKa = 3.09AHH136 pKa = 6.64GNTASGSFHH145 pKa = 6.29ITVQDD150 pKa = 3.22TTAPVLALPTPITATATSTAGAPVTFSATATDD182 pKa = 4.14IVDD185 pKa = 3.5GTDD188 pKa = 3.49PVTCTPASGSTFALGTATVTCTTSDD213 pKa = 3.29LHH215 pKa = 7.82SNTSTGSFTVTVNPGPVSTVKK236 pKa = 10.59ISPPTATTIVGSTPIQFTATSQDD259 pKa = 2.92QFGNIRR265 pKa = 11.84TGDD268 pKa = 3.52TLTWSVVSGGGSISSTGLYY287 pKa = 9.69TPGTTADD294 pKa = 3.66TATITATDD302 pKa = 4.16GSVSGTATITLNPGQVTSVTVSPSTVTLTSTQTQQFTATSQDD344 pKa = 2.92QFGNIRR350 pKa = 11.84TGDD353 pKa = 3.52TLTWSVVSGGGSISSTGLYY372 pKa = 9.7TPGTTAGTTVIAATDD387 pKa = 3.61NSISGSAIVTANNPNTIVQNQGGANLKK414 pKa = 10.22GDD416 pKa = 3.62NLQFLNLSNFNLSGDD431 pKa = 3.68NLKK434 pKa = 10.91GVTLQGSDD442 pKa = 3.46LQGANLSGANLAGANLQNVNLAGANLSGANLQGVNLSGANLQGANLKK489 pKa = 9.75GANLQGTNLQNDD501 pKa = 3.85NLLGADD507 pKa = 3.59LQGNNLQGANLSGANLQNANLSGANLQGANLQNANISGANLTGANVHH554 pKa = 6.27KK555 pKa = 10.81ANLQGTITSGITPSTLSGCLNNSVCQQ581 pKa = 3.61
MM1 pKa = 7.33TNINAKK7 pKa = 8.84ATGPSGAPVTFSATATDD24 pKa = 4.14IVDD27 pKa = 3.5GTDD30 pKa = 4.4PVTCDD35 pKa = 3.29HH36 pKa = 6.84NSGDD40 pKa = 4.16TYY42 pKa = 11.19PLGSTTVSCSSTDD55 pKa = 3.29HH56 pKa = 7.08AGNTSHH62 pKa = 7.38ASFTITVAEE71 pKa = 4.44TTAPSITLPGTITQEE86 pKa = 3.69ATGPSGAPVTFSATATDD103 pKa = 4.14IVDD106 pKa = 3.5GTDD109 pKa = 3.49PVTCTPASGSTFALGSTTVNCSSTDD134 pKa = 3.09AHH136 pKa = 6.64GNTASGSFHH145 pKa = 6.29ITVQDD150 pKa = 3.22TTAPVLALPTPITATATSTAGAPVTFSATATDD182 pKa = 4.14IVDD185 pKa = 3.5GTDD188 pKa = 3.49PVTCTPASGSTFALGTATVTCTTSDD213 pKa = 3.29LHH215 pKa = 7.82SNTSTGSFTVTVNPGPVSTVKK236 pKa = 10.59ISPPTATTIVGSTPIQFTATSQDD259 pKa = 2.92QFGNIRR265 pKa = 11.84TGDD268 pKa = 3.52TLTWSVVSGGGSISSTGLYY287 pKa = 9.69TPGTTADD294 pKa = 3.66TATITATDD302 pKa = 4.16GSVSGTATITLNPGQVTSVTVSPSTVTLTSTQTQQFTATSQDD344 pKa = 2.92QFGNIRR350 pKa = 11.84TGDD353 pKa = 3.52TLTWSVVSGGGSISSTGLYY372 pKa = 9.7TPGTTAGTTVIAATDD387 pKa = 3.61NSISGSAIVTANNPNTIVQNQGGANLKK414 pKa = 10.22GDD416 pKa = 3.62NLQFLNLSNFNLSGDD431 pKa = 3.68NLKK434 pKa = 10.91GVTLQGSDD442 pKa = 3.46LQGANLSGANLAGANLQNVNLAGANLSGANLQGVNLSGANLQGANLKK489 pKa = 9.75GANLQGTNLQNDD501 pKa = 3.85NLLGADD507 pKa = 3.59LQGNNLQGANLSGANLQNANLSGANLQGANLQNANISGANLTGANVHH554 pKa = 6.27KK555 pKa = 10.81ANLQGTITSGITPSTLSGCLNNSVCQQ581 pKa = 3.61
Molecular weight: 57.17 kDa
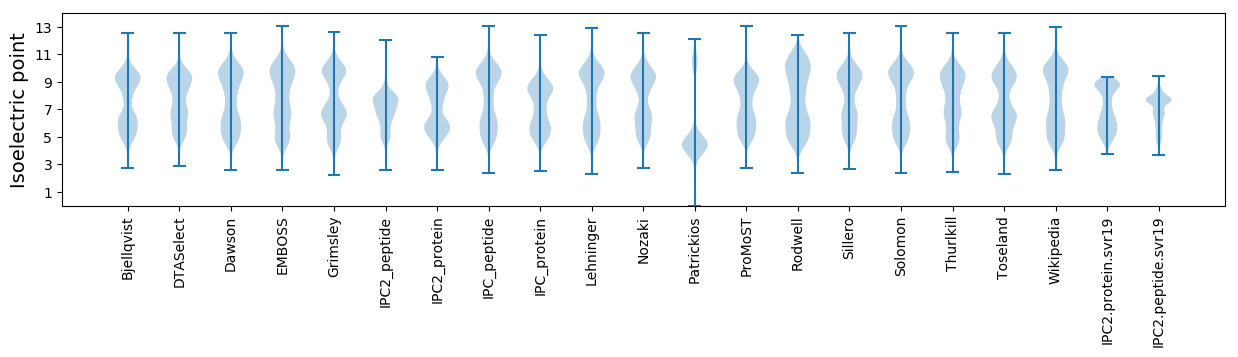
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H1EH61|A0A2H1EH61_9ARCH Uncharacterized protein OS=Candidatus Nitrosotalea sinensis OX=1499975 GN=NSIN_20732 PE=4 SV=1
MM1 pKa = 7.67AAHH4 pKa = 7.52KK5 pKa = 7.68STPRR9 pKa = 11.84KK10 pKa = 9.0NRR12 pKa = 11.84LMKK15 pKa = 10.27KK16 pKa = 9.32IKK18 pKa = 9.65QNKK21 pKa = 8.43SVPAWIIVRR30 pKa = 11.84TKK32 pKa = 10.48RR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84TNPKK40 pKa = 8.23KK41 pKa = 9.64RR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84TDD48 pKa = 3.12VKK50 pKa = 11.16VGG52 pKa = 3.24
MM1 pKa = 7.67AAHH4 pKa = 7.52KK5 pKa = 7.68STPRR9 pKa = 11.84KK10 pKa = 9.0NRR12 pKa = 11.84LMKK15 pKa = 10.27KK16 pKa = 9.32IKK18 pKa = 9.65QNKK21 pKa = 8.43SVPAWIIVRR30 pKa = 11.84TKK32 pKa = 10.48RR33 pKa = 11.84RR34 pKa = 11.84VRR36 pKa = 11.84TNPKK40 pKa = 8.23KK41 pKa = 9.64RR42 pKa = 11.84QWRR45 pKa = 11.84RR46 pKa = 11.84TDD48 pKa = 3.12VKK50 pKa = 11.16VGG52 pKa = 3.24
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
481166 |
23 |
1824 |
255.8 |
28.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.423 ± 0.06 | 0.961 ± 0.021 |
5.513 ± 0.045 | 5.781 ± 0.069 |
4.13 ± 0.04 | 6.737 ± 0.065 |
1.952 ± 0.021 | 8.552 ± 0.059 |
7.942 ± 0.077 | 8.615 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.034 | 4.778 ± 0.058 |
3.948 ± 0.044 | 3.445 ± 0.042 |
3.853 ± 0.047 | 7.498 ± 0.066 |
6.035 ± 0.073 | 6.975 ± 0.054 |
0.907 ± 0.019 | 3.249 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
