
Capybara microvirus Cap3_SP_332
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.67
Get precalculated fractions of proteins
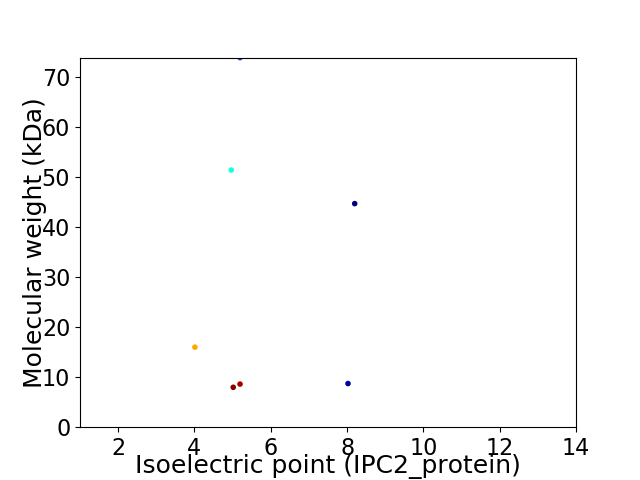
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
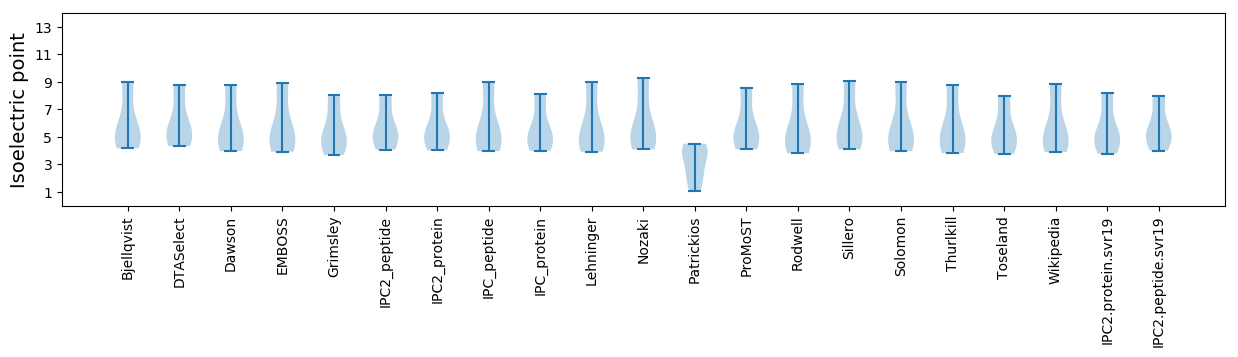
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5N2|A0A4P8W5N2_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_332 OX=2585430 PE=4 SV=1
MM1 pKa = 6.8ITKK4 pKa = 10.41NSIKK8 pKa = 10.61LSILNFLKK16 pKa = 10.8DD17 pKa = 3.59NPNFNRR23 pKa = 11.84INTLIDD29 pKa = 3.51YY30 pKa = 10.21LKK32 pKa = 10.32FDD34 pKa = 4.8PDD36 pKa = 3.87SFVHH40 pKa = 6.32SFEE43 pKa = 5.27DD44 pKa = 3.06NGVFVDD50 pKa = 5.67LKK52 pKa = 9.75FTPTSYY58 pKa = 11.46NDD60 pKa = 3.53YY61 pKa = 10.39LISALSFADD70 pKa = 3.11QVIFHH75 pKa = 6.28SFKK78 pKa = 11.14DD79 pKa = 3.59VIEE82 pKa = 4.17VVFFFEE88 pKa = 4.22PTNDD92 pKa = 3.08FKK94 pKa = 11.5FIVYY98 pKa = 9.84GIFDD102 pKa = 5.49DD103 pKa = 6.18IIPTGNTWADD113 pKa = 3.08MTFYY117 pKa = 10.69FEE119 pKa = 4.98SCIEE123 pKa = 3.79PDD125 pKa = 3.95YY126 pKa = 10.27YY127 pKa = 10.28ISMYY131 pKa = 10.67EE132 pKa = 3.91EE133 pKa = 4.42YY134 pKa = 10.39EE135 pKa = 3.98
MM1 pKa = 6.8ITKK4 pKa = 10.41NSIKK8 pKa = 10.61LSILNFLKK16 pKa = 10.8DD17 pKa = 3.59NPNFNRR23 pKa = 11.84INTLIDD29 pKa = 3.51YY30 pKa = 10.21LKK32 pKa = 10.32FDD34 pKa = 4.8PDD36 pKa = 3.87SFVHH40 pKa = 6.32SFEE43 pKa = 5.27DD44 pKa = 3.06NGVFVDD50 pKa = 5.67LKK52 pKa = 9.75FTPTSYY58 pKa = 11.46NDD60 pKa = 3.53YY61 pKa = 10.39LISALSFADD70 pKa = 3.11QVIFHH75 pKa = 6.28SFKK78 pKa = 11.14DD79 pKa = 3.59VIEE82 pKa = 4.17VVFFFEE88 pKa = 4.22PTNDD92 pKa = 3.08FKK94 pKa = 11.5FIVYY98 pKa = 9.84GIFDD102 pKa = 5.49DD103 pKa = 6.18IIPTGNTWADD113 pKa = 3.08MTFYY117 pKa = 10.69FEE119 pKa = 4.98SCIEE123 pKa = 3.79PDD125 pKa = 3.95YY126 pKa = 10.27YY127 pKa = 10.28ISMYY131 pKa = 10.67EE132 pKa = 3.91EE133 pKa = 4.42YY134 pKa = 10.39EE135 pKa = 3.98
Molecular weight: 15.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVS2|A0A4V1FVS2_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_332 OX=2585430 PE=4 SV=1
MM1 pKa = 7.16CTSPLLIKK9 pKa = 10.74NPNCLLGLHH18 pKa = 7.09DD19 pKa = 5.66GLAKK23 pKa = 10.05SLKK26 pKa = 10.57GITNTIKK33 pKa = 10.51HH34 pKa = 5.76PQSGFKK40 pKa = 10.3DD41 pKa = 3.67YY42 pKa = 10.9LSQYY46 pKa = 10.49IYY48 pKa = 10.56VPCGKK53 pKa = 10.14CPEE56 pKa = 4.52CLNSKK61 pKa = 10.42QNDD64 pKa = 3.38IIQRR68 pKa = 11.84VSFEE72 pKa = 3.91QLTSYY77 pKa = 11.1EE78 pKa = 4.4FFCTLTVKK86 pKa = 10.61PGFLDD91 pKa = 3.67SAVLPDD97 pKa = 3.55SSHH100 pKa = 6.6YY101 pKa = 11.05YY102 pKa = 9.32YY103 pKa = 11.18LDD105 pKa = 3.2WSKK108 pKa = 11.73LDD110 pKa = 3.99LFFKK114 pKa = 10.78RR115 pKa = 11.84LRR117 pKa = 11.84NKK119 pKa = 10.56NIFDD123 pKa = 4.09GPFKK127 pKa = 10.82YY128 pKa = 10.39LAVQEE133 pKa = 5.04FGHH136 pKa = 5.64EE137 pKa = 3.8THH139 pKa = 6.98RR140 pKa = 11.84PHH142 pKa = 6.41FHH144 pKa = 5.99VLFFIPRR151 pKa = 11.84LGKK154 pKa = 10.36DD155 pKa = 2.89ILWPYY160 pKa = 10.39RR161 pKa = 11.84CVDD164 pKa = 3.56EE165 pKa = 5.73NFDD168 pKa = 3.91ILSNHH173 pKa = 5.76QIKK176 pKa = 10.62SRR178 pKa = 11.84ILDD181 pKa = 3.0EE182 pKa = 4.16WYY184 pKa = 9.78INKK187 pKa = 9.68GSKK190 pKa = 10.06RR191 pKa = 11.84NPQPEE196 pKa = 4.28PLTEE200 pKa = 4.16FFTYY204 pKa = 10.1CGRR207 pKa = 11.84RR208 pKa = 11.84NFDD211 pKa = 3.34FSFVDD216 pKa = 3.72PKK218 pKa = 10.66TSQDD222 pKa = 3.57LAVDD226 pKa = 4.87FYY228 pKa = 11.62ASKK231 pKa = 11.01YY232 pKa = 9.8ALKK235 pKa = 10.47PDD237 pKa = 3.14DD238 pKa = 4.13WLNKK242 pKa = 8.97KK243 pKa = 10.35LSYY246 pKa = 10.35LLYY249 pKa = 9.61MSYY252 pKa = 11.0NFYY255 pKa = 9.59STKK258 pKa = 10.75DD259 pKa = 3.33NPYY262 pKa = 8.16YY263 pKa = 10.48LAYY266 pKa = 9.75TRR268 pKa = 11.84MRR270 pKa = 11.84PRR272 pKa = 11.84IYY274 pKa = 9.66MSHH277 pKa = 6.3QLGCSDD283 pKa = 3.09STRR286 pKa = 11.84RR287 pKa = 11.84YY288 pKa = 9.68IEE290 pKa = 5.1RR291 pKa = 11.84EE292 pKa = 3.82TLDD295 pKa = 4.57NICLNYY301 pKa = 9.39PYY303 pKa = 10.28PVIHH307 pKa = 6.62SPADD311 pKa = 3.15GRR313 pKa = 11.84TYY315 pKa = 10.6PLSAYY320 pKa = 9.22FRR322 pKa = 11.84KK323 pKa = 10.15KK324 pKa = 10.0YY325 pKa = 10.69LPARR329 pKa = 11.84WYY331 pKa = 10.51DD332 pKa = 4.19DD333 pKa = 3.06MLLYY337 pKa = 9.84YY338 pKa = 10.15QRR340 pKa = 11.84NNMPLATFTHH350 pKa = 6.54NEE352 pKa = 3.88DD353 pKa = 2.96KK354 pKa = 10.98HH355 pKa = 5.84RR356 pKa = 11.84QNAINFDD363 pKa = 4.07KK364 pKa = 10.67KK365 pKa = 10.34CKK367 pKa = 10.1KK368 pKa = 9.93ILEE371 pKa = 4.11HH372 pKa = 7.19HH373 pKa = 6.48ILL375 pKa = 3.76
MM1 pKa = 7.16CTSPLLIKK9 pKa = 10.74NPNCLLGLHH18 pKa = 7.09DD19 pKa = 5.66GLAKK23 pKa = 10.05SLKK26 pKa = 10.57GITNTIKK33 pKa = 10.51HH34 pKa = 5.76PQSGFKK40 pKa = 10.3DD41 pKa = 3.67YY42 pKa = 10.9LSQYY46 pKa = 10.49IYY48 pKa = 10.56VPCGKK53 pKa = 10.14CPEE56 pKa = 4.52CLNSKK61 pKa = 10.42QNDD64 pKa = 3.38IIQRR68 pKa = 11.84VSFEE72 pKa = 3.91QLTSYY77 pKa = 11.1EE78 pKa = 4.4FFCTLTVKK86 pKa = 10.61PGFLDD91 pKa = 3.67SAVLPDD97 pKa = 3.55SSHH100 pKa = 6.6YY101 pKa = 11.05YY102 pKa = 9.32YY103 pKa = 11.18LDD105 pKa = 3.2WSKK108 pKa = 11.73LDD110 pKa = 3.99LFFKK114 pKa = 10.78RR115 pKa = 11.84LRR117 pKa = 11.84NKK119 pKa = 10.56NIFDD123 pKa = 4.09GPFKK127 pKa = 10.82YY128 pKa = 10.39LAVQEE133 pKa = 5.04FGHH136 pKa = 5.64EE137 pKa = 3.8THH139 pKa = 6.98RR140 pKa = 11.84PHH142 pKa = 6.41FHH144 pKa = 5.99VLFFIPRR151 pKa = 11.84LGKK154 pKa = 10.36DD155 pKa = 2.89ILWPYY160 pKa = 10.39RR161 pKa = 11.84CVDD164 pKa = 3.56EE165 pKa = 5.73NFDD168 pKa = 3.91ILSNHH173 pKa = 5.76QIKK176 pKa = 10.62SRR178 pKa = 11.84ILDD181 pKa = 3.0EE182 pKa = 4.16WYY184 pKa = 9.78INKK187 pKa = 9.68GSKK190 pKa = 10.06RR191 pKa = 11.84NPQPEE196 pKa = 4.28PLTEE200 pKa = 4.16FFTYY204 pKa = 10.1CGRR207 pKa = 11.84RR208 pKa = 11.84NFDD211 pKa = 3.34FSFVDD216 pKa = 3.72PKK218 pKa = 10.66TSQDD222 pKa = 3.57LAVDD226 pKa = 4.87FYY228 pKa = 11.62ASKK231 pKa = 11.01YY232 pKa = 9.8ALKK235 pKa = 10.47PDD237 pKa = 3.14DD238 pKa = 4.13WLNKK242 pKa = 8.97KK243 pKa = 10.35LSYY246 pKa = 10.35LLYY249 pKa = 9.61MSYY252 pKa = 11.0NFYY255 pKa = 9.59STKK258 pKa = 10.75DD259 pKa = 3.33NPYY262 pKa = 8.16YY263 pKa = 10.48LAYY266 pKa = 9.75TRR268 pKa = 11.84MRR270 pKa = 11.84PRR272 pKa = 11.84IYY274 pKa = 9.66MSHH277 pKa = 6.3QLGCSDD283 pKa = 3.09STRR286 pKa = 11.84RR287 pKa = 11.84YY288 pKa = 9.68IEE290 pKa = 5.1RR291 pKa = 11.84EE292 pKa = 3.82TLDD295 pKa = 4.57NICLNYY301 pKa = 9.39PYY303 pKa = 10.28PVIHH307 pKa = 6.62SPADD311 pKa = 3.15GRR313 pKa = 11.84TYY315 pKa = 10.6PLSAYY320 pKa = 9.22FRR322 pKa = 11.84KK323 pKa = 10.15KK324 pKa = 10.0YY325 pKa = 10.69LPARR329 pKa = 11.84WYY331 pKa = 10.51DD332 pKa = 4.19DD333 pKa = 3.06MLLYY337 pKa = 9.84YY338 pKa = 10.15QRR340 pKa = 11.84NNMPLATFTHH350 pKa = 6.54NEE352 pKa = 3.88DD353 pKa = 2.96KK354 pKa = 10.98HH355 pKa = 5.84RR356 pKa = 11.84QNAINFDD363 pKa = 4.07KK364 pKa = 10.67KK365 pKa = 10.34CKK367 pKa = 10.1KK368 pKa = 9.93ILEE371 pKa = 4.11HH372 pKa = 7.19HH373 pKa = 6.48ILL375 pKa = 3.76
Molecular weight: 44.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1834 |
71 |
640 |
262.0 |
30.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.344 ± 1.353 | 1.527 ± 0.519 |
7.361 ± 0.39 | 3.98 ± 0.523 |
6.543 ± 0.873 | 4.526 ± 0.735 |
1.854 ± 0.524 | 6.052 ± 0.917 |
5.616 ± 0.564 | 8.342 ± 0.596 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.617 ± 0.35 | 7.306 ± 0.418 |
4.253 ± 0.665 | 3.762 ± 1.135 |
4.144 ± 0.473 | 8.724 ± 0.604 |
5.125 ± 0.571 | 5.507 ± 1.194 |
1.2 ± 0.161 | 6.216 ± 0.813 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
