
Monosporascus sp. GIB2
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Xylariales incertae sedis; Monosporascus; unclassified Monosporascus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
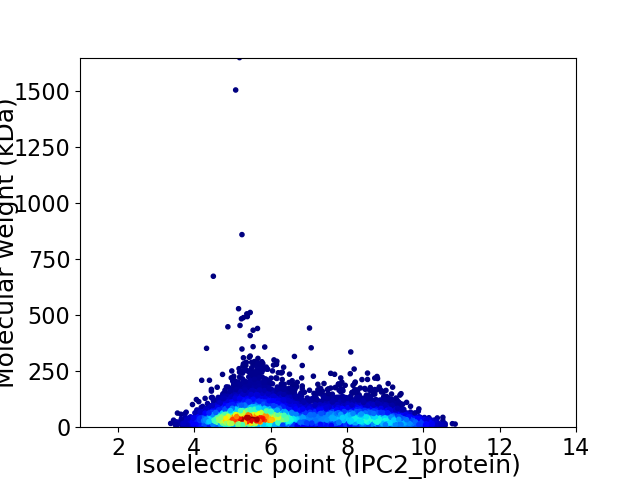
Virtual 2D-PAGE plot for 11773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q4TPP1|A0A4Q4TPP1_9PEZI Uncharacterized protein OS=Monosporascus sp. GIB2 OX=2211647 GN=DL765_009128 PE=4 SV=1
MM1 pKa = 7.1NLCMAIAAPRR11 pKa = 11.84SQDD14 pKa = 3.08GFFSLPASRR23 pKa = 11.84ISKK26 pKa = 9.68PLTVLKK32 pKa = 10.33RR33 pKa = 11.84QEE35 pKa = 4.15DD36 pKa = 4.28TDD38 pKa = 3.84VPLYY42 pKa = 10.65DD43 pKa = 3.55VSEE46 pKa = 4.07VSYY49 pKa = 10.81LIEE52 pKa = 4.63LSIGTPGQSVKK63 pKa = 10.64VAIDD67 pKa = 3.45TGSSEE72 pKa = 4.36LWVDD76 pKa = 3.91PVCQNSQSLSEE87 pKa = 4.08VDD89 pKa = 3.2EE90 pKa = 5.26CIANGIYY97 pKa = 10.41DD98 pKa = 4.39PSQSTTFEE106 pKa = 4.63DD107 pKa = 3.97VNSTNTIPYY116 pKa = 8.48GIGVVRR122 pKa = 11.84IEE124 pKa = 3.94YY125 pKa = 10.39VRR127 pKa = 11.84DD128 pKa = 3.23NVAFPNSTINLTDD141 pKa = 3.25VQFGVATASRR151 pKa = 11.84QLNEE155 pKa = 4.58GIMGLSFGGGTPEE168 pKa = 5.07ADD170 pKa = 2.85LRR172 pKa = 11.84YY173 pKa = 10.62NNIVDD178 pKa = 3.99EE179 pKa = 5.43LYY181 pKa = 10.69LQNVTQSRR189 pKa = 11.84AFSVALGSSDD199 pKa = 3.6SDD201 pKa = 3.46NDD203 pKa = 3.72SVVIFGGIDD212 pKa = 3.13TSKK215 pKa = 10.49YY216 pKa = 10.15AGPLRR221 pKa = 11.84TLPILGPQNGEE232 pKa = 3.74TLNRR236 pKa = 11.84YY237 pKa = 7.23WVQLDD242 pKa = 4.19SISTTVDD249 pKa = 3.43GSSHH253 pKa = 6.46TYY255 pKa = 10.38DD256 pKa = 3.31SSSLPVVLDD265 pKa = 3.52TGSTFCSLPPAVVNGMMEE283 pKa = 4.22DD284 pKa = 4.0LGGQINNQGQVLVDD298 pKa = 4.02CSLVDD303 pKa = 3.75NEE305 pKa = 4.66GTFDD309 pKa = 4.06FDD311 pKa = 4.47FGNITIRR318 pKa = 11.84IPYY321 pKa = 8.46SQFVIQANSEE331 pKa = 4.39VCVLGATPDD340 pKa = 3.67DD341 pKa = 4.25QIALLGDD348 pKa = 3.49SFLRR352 pKa = 11.84SAYY355 pKa = 10.54VVFDD359 pKa = 3.77QNNMEE364 pKa = 4.36VAMAPYY370 pKa = 9.87ANCGQTEE377 pKa = 3.71QRR379 pKa = 11.84IPDD382 pKa = 3.87GGVSGVKK389 pKa = 10.04GAEE392 pKa = 3.86QTATDD397 pKa = 3.95IDD399 pKa = 4.16GDD401 pKa = 4.41DD402 pKa = 3.53NVMLGG407 pKa = 3.53
MM1 pKa = 7.1NLCMAIAAPRR11 pKa = 11.84SQDD14 pKa = 3.08GFFSLPASRR23 pKa = 11.84ISKK26 pKa = 9.68PLTVLKK32 pKa = 10.33RR33 pKa = 11.84QEE35 pKa = 4.15DD36 pKa = 4.28TDD38 pKa = 3.84VPLYY42 pKa = 10.65DD43 pKa = 3.55VSEE46 pKa = 4.07VSYY49 pKa = 10.81LIEE52 pKa = 4.63LSIGTPGQSVKK63 pKa = 10.64VAIDD67 pKa = 3.45TGSSEE72 pKa = 4.36LWVDD76 pKa = 3.91PVCQNSQSLSEE87 pKa = 4.08VDD89 pKa = 3.2EE90 pKa = 5.26CIANGIYY97 pKa = 10.41DD98 pKa = 4.39PSQSTTFEE106 pKa = 4.63DD107 pKa = 3.97VNSTNTIPYY116 pKa = 8.48GIGVVRR122 pKa = 11.84IEE124 pKa = 3.94YY125 pKa = 10.39VRR127 pKa = 11.84DD128 pKa = 3.23NVAFPNSTINLTDD141 pKa = 3.25VQFGVATASRR151 pKa = 11.84QLNEE155 pKa = 4.58GIMGLSFGGGTPEE168 pKa = 5.07ADD170 pKa = 2.85LRR172 pKa = 11.84YY173 pKa = 10.62NNIVDD178 pKa = 3.99EE179 pKa = 5.43LYY181 pKa = 10.69LQNVTQSRR189 pKa = 11.84AFSVALGSSDD199 pKa = 3.6SDD201 pKa = 3.46NDD203 pKa = 3.72SVVIFGGIDD212 pKa = 3.13TSKK215 pKa = 10.49YY216 pKa = 10.15AGPLRR221 pKa = 11.84TLPILGPQNGEE232 pKa = 3.74TLNRR236 pKa = 11.84YY237 pKa = 7.23WVQLDD242 pKa = 4.19SISTTVDD249 pKa = 3.43GSSHH253 pKa = 6.46TYY255 pKa = 10.38DD256 pKa = 3.31SSSLPVVLDD265 pKa = 3.52TGSTFCSLPPAVVNGMMEE283 pKa = 4.22DD284 pKa = 4.0LGGQINNQGQVLVDD298 pKa = 4.02CSLVDD303 pKa = 3.75NEE305 pKa = 4.66GTFDD309 pKa = 4.06FDD311 pKa = 4.47FGNITIRR318 pKa = 11.84IPYY321 pKa = 8.46SQFVIQANSEE331 pKa = 4.39VCVLGATPDD340 pKa = 3.67DD341 pKa = 4.25QIALLGDD348 pKa = 3.49SFLRR352 pKa = 11.84SAYY355 pKa = 10.54VVFDD359 pKa = 3.77QNNMEE364 pKa = 4.36VAMAPYY370 pKa = 9.87ANCGQTEE377 pKa = 3.71QRR379 pKa = 11.84IPDD382 pKa = 3.87GGVSGVKK389 pKa = 10.04GAEE392 pKa = 3.86QTATDD397 pKa = 3.95IDD399 pKa = 4.16GDD401 pKa = 4.41DD402 pKa = 3.53NVMLGG407 pKa = 3.53
Molecular weight: 43.57 kDa
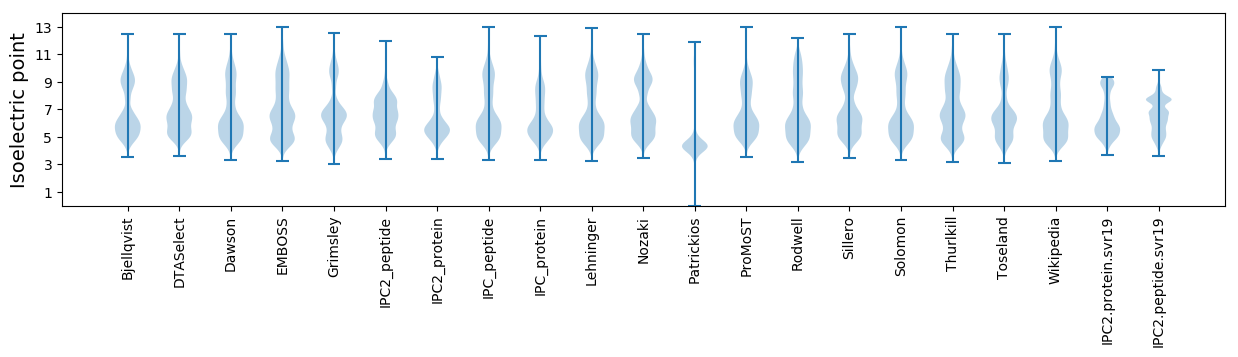
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q4TZB6|A0A4Q4TZB6_9PEZI Uncharacterized protein OS=Monosporascus sp. GIB2 OX=2211647 GN=DL765_007306 PE=4 SV=1
MM1 pKa = 7.61ASAGAWEE8 pKa = 4.62ARR10 pKa = 11.84TPPLAATSAMVLTAMLSEE28 pKa = 4.41SAMSVTGAVNATGAISTIIRR48 pKa = 11.84CGGYY52 pKa = 9.38SGEE55 pKa = 4.36VGCVPRR61 pKa = 11.84GAAVDD66 pKa = 4.04LGSAGRR72 pKa = 11.84IDD74 pKa = 3.18ITGAVIVPSVLLLLPPANKK93 pKa = 10.01SVGHH97 pKa = 5.23VRR99 pKa = 11.84GPQPDD104 pKa = 3.72VTHH107 pKa = 6.79KK108 pKa = 10.86LVLVSIASKK117 pKa = 9.9TPMAKK122 pKa = 9.47PARR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84TWSMMKK133 pKa = 9.91AQSDD137 pKa = 3.95AAPVRR142 pKa = 11.84LDD144 pKa = 3.75DD145 pKa = 4.16NVGKK149 pKa = 10.5GLEE152 pKa = 4.21PLHH155 pKa = 6.01HH156 pKa = 6.78HH157 pKa = 7.1RR158 pKa = 11.84LPVAYY163 pKa = 9.55FSALIDD169 pKa = 3.28RR170 pKa = 11.84LFARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84CGMLCDD182 pKa = 4.13SRR184 pKa = 11.84FAMHH188 pKa = 7.05LSASLSSNPNALLHH202 pKa = 6.14EE203 pKa = 4.4MRR205 pKa = 11.84QRR207 pKa = 11.84HH208 pKa = 5.74AAGQPVGCSSTRR220 pKa = 11.84HH221 pKa = 4.91FGRR224 pKa = 11.84RR225 pKa = 11.84YY226 pKa = 9.83GLLVNDD232 pKa = 4.83RR233 pKa = 11.84AAMAALYY240 pKa = 10.15HH241 pKa = 6.11SRR243 pKa = 3.83
MM1 pKa = 7.61ASAGAWEE8 pKa = 4.62ARR10 pKa = 11.84TPPLAATSAMVLTAMLSEE28 pKa = 4.41SAMSVTGAVNATGAISTIIRR48 pKa = 11.84CGGYY52 pKa = 9.38SGEE55 pKa = 4.36VGCVPRR61 pKa = 11.84GAAVDD66 pKa = 4.04LGSAGRR72 pKa = 11.84IDD74 pKa = 3.18ITGAVIVPSVLLLLPPANKK93 pKa = 10.01SVGHH97 pKa = 5.23VRR99 pKa = 11.84GPQPDD104 pKa = 3.72VTHH107 pKa = 6.79KK108 pKa = 10.86LVLVSIASKK117 pKa = 9.9TPMAKK122 pKa = 9.47PARR125 pKa = 11.84RR126 pKa = 11.84RR127 pKa = 11.84TWSMMKK133 pKa = 9.91AQSDD137 pKa = 3.95AAPVRR142 pKa = 11.84LDD144 pKa = 3.75DD145 pKa = 4.16NVGKK149 pKa = 10.5GLEE152 pKa = 4.21PLHH155 pKa = 6.01HH156 pKa = 6.78HH157 pKa = 7.1RR158 pKa = 11.84LPVAYY163 pKa = 9.55FSALIDD169 pKa = 3.28RR170 pKa = 11.84LFARR174 pKa = 11.84RR175 pKa = 11.84RR176 pKa = 11.84CGMLCDD182 pKa = 4.13SRR184 pKa = 11.84FAMHH188 pKa = 7.05LSASLSSNPNALLHH202 pKa = 6.14EE203 pKa = 4.4MRR205 pKa = 11.84QRR207 pKa = 11.84HH208 pKa = 5.74AAGQPVGCSSTRR220 pKa = 11.84HH221 pKa = 4.91FGRR224 pKa = 11.84RR225 pKa = 11.84YY226 pKa = 9.83GLLVNDD232 pKa = 4.83RR233 pKa = 11.84AAMAALYY240 pKa = 10.15HH241 pKa = 6.11SRR243 pKa = 3.83
Molecular weight: 25.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5809346 |
66 |
14987 |
493.4 |
54.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.278 ± 0.021 | 1.189 ± 0.009 |
5.905 ± 0.015 | 6.318 ± 0.023 |
3.532 ± 0.012 | 7.344 ± 0.023 |
2.339 ± 0.009 | 4.522 ± 0.016 |
4.572 ± 0.021 | 8.761 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.097 ± 0.008 | 3.487 ± 0.012 |
6.211 ± 0.023 | 3.854 ± 0.015 |
6.627 ± 0.02 | 7.856 ± 0.021 |
5.733 ± 0.016 | 6.198 ± 0.018 |
1.469 ± 0.007 | 2.709 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
