
Cycloclasticus sp. symbiont of Poecilosclerida sp. N
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Cycloclasticus; unclassified Cycloclasticus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
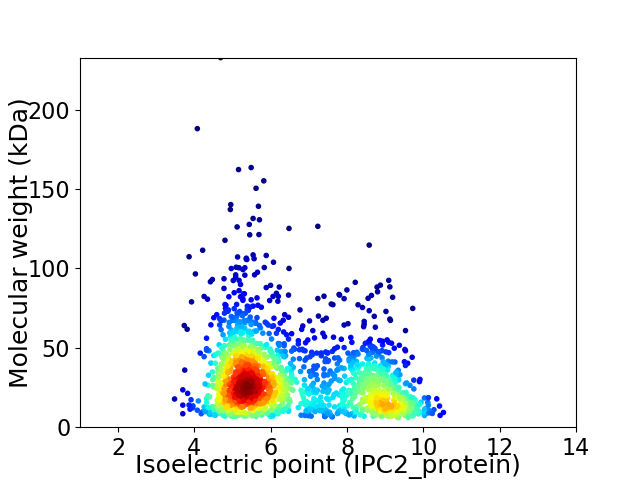
Virtual 2D-PAGE plot for 1736 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X1QJF7|A0A1X1QJF7_9GAMM Uncharacterized protein OS=Cycloclasticus sp. symbiont of Poecilosclerida sp. N OX=1840472 GN=A6F72_01500 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.09LLKK5 pKa = 9.19ITAIASLLAASGAASAVEE23 pKa = 3.78ISGNVALTTDD33 pKa = 4.0YY34 pKa = 10.66VWRR37 pKa = 11.84GISQTDD43 pKa = 3.9GSPAIQGGFDD53 pKa = 3.04AGFGNGLYY61 pKa = 10.02TGIWASNVDD70 pKa = 4.05FGGDD74 pKa = 3.01EE75 pKa = 4.31SMEE78 pKa = 4.39LDD80 pKa = 4.91VYY82 pKa = 11.21AGWAGEE88 pKa = 4.08FKK90 pKa = 10.92SVGVDD95 pKa = 2.84IGVLHH100 pKa = 5.92YY101 pKa = 10.13AYY103 pKa = 7.2PTSAPDD109 pKa = 3.38TDD111 pKa = 3.92FTEE114 pKa = 5.55AYY116 pKa = 9.88AGLSYY121 pKa = 11.14GPASLTQYY129 pKa = 10.73FGFDD133 pKa = 3.66AGSDD137 pKa = 3.9DD138 pKa = 4.71IGDD141 pKa = 3.65VDD143 pKa = 4.56FGNYY147 pKa = 7.87TDD149 pKa = 5.18LGLDD153 pKa = 3.14LGEE156 pKa = 4.32YY157 pKa = 10.46NGITLAVHH165 pKa = 6.77AGHH168 pKa = 7.08YY169 pKa = 9.25DD170 pKa = 3.45RR171 pKa = 11.84KK172 pKa = 10.59SGSDD176 pKa = 4.56DD177 pKa = 3.26YY178 pKa = 11.54WDD180 pKa = 3.26WRR182 pKa = 11.84LAASTSLFGVDD193 pKa = 3.08WEE195 pKa = 4.57VAYY198 pKa = 11.2SDD200 pKa = 3.8VDD202 pKa = 3.47NANEE206 pKa = 5.11DD207 pKa = 4.03RR208 pKa = 11.84NDD210 pKa = 3.77DD211 pKa = 3.98PNLVLTVSKK220 pKa = 10.98SLL222 pKa = 3.56
MM1 pKa = 7.63KK2 pKa = 10.09LLKK5 pKa = 9.19ITAIASLLAASGAASAVEE23 pKa = 3.78ISGNVALTTDD33 pKa = 4.0YY34 pKa = 10.66VWRR37 pKa = 11.84GISQTDD43 pKa = 3.9GSPAIQGGFDD53 pKa = 3.04AGFGNGLYY61 pKa = 10.02TGIWASNVDD70 pKa = 4.05FGGDD74 pKa = 3.01EE75 pKa = 4.31SMEE78 pKa = 4.39LDD80 pKa = 4.91VYY82 pKa = 11.21AGWAGEE88 pKa = 4.08FKK90 pKa = 10.92SVGVDD95 pKa = 2.84IGVLHH100 pKa = 5.92YY101 pKa = 10.13AYY103 pKa = 7.2PTSAPDD109 pKa = 3.38TDD111 pKa = 3.92FTEE114 pKa = 5.55AYY116 pKa = 9.88AGLSYY121 pKa = 11.14GPASLTQYY129 pKa = 10.73FGFDD133 pKa = 3.66AGSDD137 pKa = 3.9DD138 pKa = 4.71IGDD141 pKa = 3.65VDD143 pKa = 4.56FGNYY147 pKa = 7.87TDD149 pKa = 5.18LGLDD153 pKa = 3.14LGEE156 pKa = 4.32YY157 pKa = 10.46NGITLAVHH165 pKa = 6.77AGHH168 pKa = 7.08YY169 pKa = 9.25DD170 pKa = 3.45RR171 pKa = 11.84KK172 pKa = 10.59SGSDD176 pKa = 4.56DD177 pKa = 3.26YY178 pKa = 11.54WDD180 pKa = 3.26WRR182 pKa = 11.84LAASTSLFGVDD193 pKa = 3.08WEE195 pKa = 4.57VAYY198 pKa = 11.2SDD200 pKa = 3.8VDD202 pKa = 3.47NANEE206 pKa = 5.11DD207 pKa = 4.03RR208 pKa = 11.84NDD210 pKa = 3.77DD211 pKa = 3.98PNLVLTVSKK220 pKa = 10.98SLL222 pKa = 3.56
Molecular weight: 23.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X1QMH6|A0A1X1QMH6_9GAMM Nitrate reductase catalytic subunit OS=Cycloclasticus sp. symbiont of Poecilosclerida sp. N OX=1840472 GN=A6F72_04800 PE=3 SV=1
MM1 pKa = 7.43ARR3 pKa = 11.84IAGINIPDD11 pKa = 3.76NKK13 pKa = 10.35HH14 pKa = 5.44IVISLTSIYY23 pKa = 10.69GVGPTRR29 pKa = 11.84SKK31 pKa = 10.95AICKK35 pKa = 9.59AAAIDD40 pKa = 4.0TATKK44 pKa = 10.15ICDD47 pKa = 3.51LSEE50 pKa = 4.49DD51 pKa = 3.54QLEE54 pKa = 4.29LVRR57 pKa = 11.84TEE59 pKa = 3.63VGKK62 pKa = 10.69YY63 pKa = 9.88IIEE66 pKa = 4.58GDD68 pKa = 3.49LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.4VSMNIKK78 pKa = 10.26RR79 pKa = 11.84LLDD82 pKa = 3.94LGCFRR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 5.94RR92 pKa = 11.84RR93 pKa = 11.84SLPVRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 10.45TNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.72GPRR113 pKa = 11.84KK114 pKa = 9.36PIRR117 pKa = 11.84KK118 pKa = 9.01
MM1 pKa = 7.43ARR3 pKa = 11.84IAGINIPDD11 pKa = 3.76NKK13 pKa = 10.35HH14 pKa = 5.44IVISLTSIYY23 pKa = 10.69GVGPTRR29 pKa = 11.84SKK31 pKa = 10.95AICKK35 pKa = 9.59AAAIDD40 pKa = 4.0TATKK44 pKa = 10.15ICDD47 pKa = 3.51LSEE50 pKa = 4.49DD51 pKa = 3.54QLEE54 pKa = 4.29LVRR57 pKa = 11.84TEE59 pKa = 3.63VGKK62 pKa = 10.69YY63 pKa = 9.88IIEE66 pKa = 4.58GDD68 pKa = 3.49LRR70 pKa = 11.84RR71 pKa = 11.84EE72 pKa = 3.4VSMNIKK78 pKa = 10.26RR79 pKa = 11.84LLDD82 pKa = 3.94LGCFRR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 5.94RR92 pKa = 11.84RR93 pKa = 11.84SLPVRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 10.45TNARR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.72GPRR113 pKa = 11.84KK114 pKa = 9.36PIRR117 pKa = 11.84KK118 pKa = 9.01
Molecular weight: 13.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
513302 |
51 |
2124 |
295.7 |
32.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.399 ± 0.069 | 1.089 ± 0.023 |
5.628 ± 0.04 | 6.206 ± 0.059 |
4.068 ± 0.04 | 7.058 ± 0.057 |
2.187 ± 0.03 | 6.71 ± 0.044 |
6.317 ± 0.066 | 10.169 ± 0.077 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.534 ± 0.032 | 4.38 ± 0.051 |
3.759 ± 0.039 | 4.054 ± 0.044 |
4.672 ± 0.039 | 6.55 ± 0.05 |
5.356 ± 0.058 | 6.911 ± 0.051 |
1.105 ± 0.021 | 2.846 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
