
Xanthobacter autotrophicus (strain ATCC BAA-1158 / Py2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Xanthobacteraceae; Xanthobacter; Xanthobacter autotrophicus
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
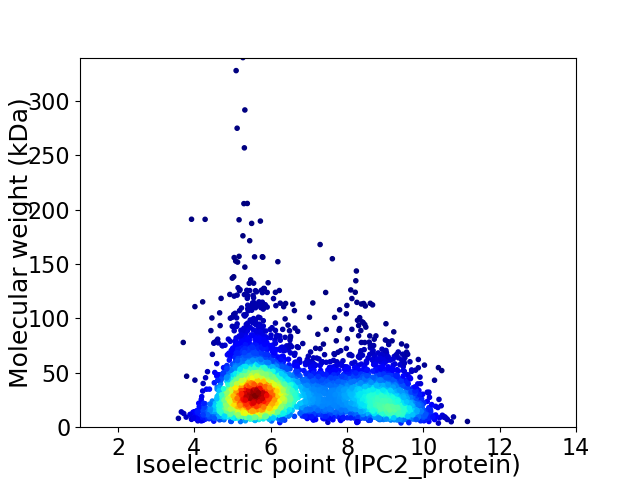
Virtual 2D-PAGE plot for 4971 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
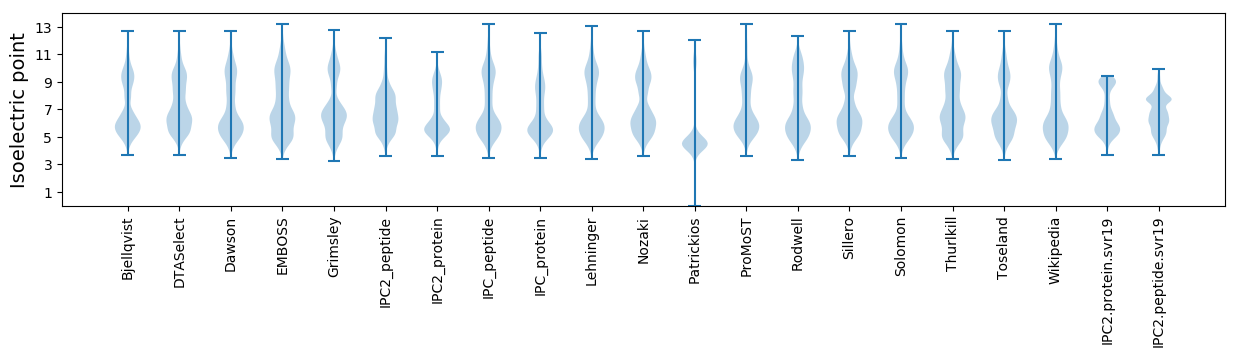
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7IDR0|A7IDR0_XANP2 Phenylacetic acid degradation protein PaaD OS=Xanthobacter autotrophicus (strain ATCC BAA-1158 / Py2) OX=78245 GN=Xaut_0902 PE=4 SV=1
MM1 pKa = 7.28ATRR4 pKa = 11.84TITWAWGAQEE14 pKa = 4.68VVSFDD19 pKa = 3.63PATDD23 pKa = 3.38VLDD26 pKa = 4.49FGWLNASDD34 pKa = 3.82FTITEE39 pKa = 4.2VNGSVVIALPANAHH53 pKa = 6.18SYY55 pKa = 8.3TLEE58 pKa = 4.24GVSLSDD64 pKa = 4.49LSLANISAFDD74 pKa = 3.75ASVFAQWSAAIAADD88 pKa = 3.94GATTPVAGTLFALSAARR105 pKa = 11.84DD106 pKa = 3.66TDD108 pKa = 3.89TVLSFNPAADD118 pKa = 3.68KK119 pKa = 11.34LDD121 pKa = 3.74FSGLSASDD129 pKa = 3.47FSIAEE134 pKa = 4.11VNGSVVIYY142 pKa = 9.8LSAEE146 pKa = 3.78RR147 pKa = 11.84QTYY150 pKa = 9.99ILDD153 pKa = 3.92GVTLSQLSLSSIASQDD169 pKa = 3.46AAVLAEE175 pKa = 4.55WSQALASPAGSSAAPTSGGGEE196 pKa = 3.9AVTTTLGWNWNTQTVLAFDD215 pKa = 4.25PARR218 pKa = 11.84DD219 pKa = 3.82KK220 pKa = 11.22IDD222 pKa = 4.18FGWLTPQDD230 pKa = 3.84FSVVEE235 pKa = 4.11LNGSVVILMPGAQQSYY251 pKa = 7.31TLEE254 pKa = 4.28GVTLAEE260 pKa = 4.38LSAANITARR269 pKa = 11.84SAVVLEE275 pKa = 4.57EE276 pKa = 3.85WATLLSGASGSGGGSGGGSGSTLPVIEE303 pKa = 4.1ASAWIDD309 pKa = 3.09AKK311 pKa = 11.18AYY313 pKa = 9.84VAGDD317 pKa = 3.88LVSVGHH323 pKa = 7.11LVYY326 pKa = 10.31QANWWTLGTDD336 pKa = 3.37PTSDD340 pKa = 3.12HH341 pKa = 6.31GAVGTGHH348 pKa = 5.59VWSVVGYY355 pKa = 11.29ADD357 pKa = 4.37LTPVAPDD364 pKa = 3.75APDD367 pKa = 3.88GLHH370 pKa = 7.13LLTTTEE376 pKa = 4.17TSATLTWDD384 pKa = 3.17AAEE387 pKa = 4.07VSGVGTVTGYY397 pKa = 11.16AIYY400 pKa = 10.22RR401 pKa = 11.84DD402 pKa = 3.98GEE404 pKa = 4.49LVGTTTDD411 pKa = 3.3LSFKK415 pKa = 10.76VSGLVADD422 pKa = 4.25TTYY425 pKa = 10.56HH426 pKa = 6.1FSVVAVDD433 pKa = 3.75EE434 pKa = 5.22AGTSPAATPIAVTTDD449 pKa = 3.07APGTDD454 pKa = 4.02AADD457 pKa = 3.53QQHH460 pKa = 6.18FSPYY464 pKa = 10.04FEE466 pKa = 4.05MWLPSSQNLVQTVEE480 pKa = 4.4DD481 pKa = 3.67VGLTAVTLAFVLGTGPDD498 pKa = 3.45QIGWGGLGSIDD509 pKa = 5.26NDD511 pKa = 3.64TLANGTTISSMVAALQQNGVEE532 pKa = 4.33VTISFGGGYY541 pKa = 8.99GQEE544 pKa = 4.11PALSFTNVAQLTAAYY559 pKa = 9.58QSVMDD564 pKa = 4.61KK565 pKa = 11.47YY566 pKa = 11.44NVTSLDD572 pKa = 3.38FDD574 pKa = 3.92IEE576 pKa = 4.41ADD578 pKa = 3.48ALTNTAASHH587 pKa = 6.1LRR589 pKa = 11.84NEE591 pKa = 4.29ALVALEE597 pKa = 4.1QANPDD602 pKa = 3.51LTVSFTLPALPTGLNQAGLDD622 pKa = 3.72LLAQAKK628 pKa = 10.26ADD630 pKa = 3.49GVEE633 pKa = 3.87IDD635 pKa = 3.95TVNIMVMNYY644 pKa = 9.07GAYY647 pKa = 9.93YY648 pKa = 10.69DD649 pKa = 4.12SGDD652 pKa = 3.68MGKK655 pKa = 10.07DD656 pKa = 3.15AIDD659 pKa = 3.58AAEE662 pKa = 3.8ATIAQLHH669 pKa = 5.07QLGLDD674 pKa = 3.35AKK676 pKa = 10.46VAITPMIGQNDD687 pKa = 3.57VPGEE691 pKa = 4.22VFTLDD696 pKa = 4.66DD697 pKa = 3.85ARR699 pKa = 11.84QLLDD703 pKa = 3.41YY704 pKa = 11.56AEE706 pKa = 4.92GNDD709 pKa = 4.05HH710 pKa = 6.81IAYY713 pKa = 9.35IAMWSLGRR721 pKa = 11.84DD722 pKa = 3.07HH723 pKa = 7.29GDD725 pKa = 3.38DD726 pKa = 3.55VGHH729 pKa = 6.37LTEE732 pKa = 4.57TSSGVAQHH740 pKa = 7.29DD741 pKa = 3.83YY742 pKa = 11.47DD743 pKa = 3.73FAKK746 pKa = 10.12IFSMVV751 pKa = 2.75
MM1 pKa = 7.28ATRR4 pKa = 11.84TITWAWGAQEE14 pKa = 4.68VVSFDD19 pKa = 3.63PATDD23 pKa = 3.38VLDD26 pKa = 4.49FGWLNASDD34 pKa = 3.82FTITEE39 pKa = 4.2VNGSVVIALPANAHH53 pKa = 6.18SYY55 pKa = 8.3TLEE58 pKa = 4.24GVSLSDD64 pKa = 4.49LSLANISAFDD74 pKa = 3.75ASVFAQWSAAIAADD88 pKa = 3.94GATTPVAGTLFALSAARR105 pKa = 11.84DD106 pKa = 3.66TDD108 pKa = 3.89TVLSFNPAADD118 pKa = 3.68KK119 pKa = 11.34LDD121 pKa = 3.74FSGLSASDD129 pKa = 3.47FSIAEE134 pKa = 4.11VNGSVVIYY142 pKa = 9.8LSAEE146 pKa = 3.78RR147 pKa = 11.84QTYY150 pKa = 9.99ILDD153 pKa = 3.92GVTLSQLSLSSIASQDD169 pKa = 3.46AAVLAEE175 pKa = 4.55WSQALASPAGSSAAPTSGGGEE196 pKa = 3.9AVTTTLGWNWNTQTVLAFDD215 pKa = 4.25PARR218 pKa = 11.84DD219 pKa = 3.82KK220 pKa = 11.22IDD222 pKa = 4.18FGWLTPQDD230 pKa = 3.84FSVVEE235 pKa = 4.11LNGSVVILMPGAQQSYY251 pKa = 7.31TLEE254 pKa = 4.28GVTLAEE260 pKa = 4.38LSAANITARR269 pKa = 11.84SAVVLEE275 pKa = 4.57EE276 pKa = 3.85WATLLSGASGSGGGSGGGSGSTLPVIEE303 pKa = 4.1ASAWIDD309 pKa = 3.09AKK311 pKa = 11.18AYY313 pKa = 9.84VAGDD317 pKa = 3.88LVSVGHH323 pKa = 7.11LVYY326 pKa = 10.31QANWWTLGTDD336 pKa = 3.37PTSDD340 pKa = 3.12HH341 pKa = 6.31GAVGTGHH348 pKa = 5.59VWSVVGYY355 pKa = 11.29ADD357 pKa = 4.37LTPVAPDD364 pKa = 3.75APDD367 pKa = 3.88GLHH370 pKa = 7.13LLTTTEE376 pKa = 4.17TSATLTWDD384 pKa = 3.17AAEE387 pKa = 4.07VSGVGTVTGYY397 pKa = 11.16AIYY400 pKa = 10.22RR401 pKa = 11.84DD402 pKa = 3.98GEE404 pKa = 4.49LVGTTTDD411 pKa = 3.3LSFKK415 pKa = 10.76VSGLVADD422 pKa = 4.25TTYY425 pKa = 10.56HH426 pKa = 6.1FSVVAVDD433 pKa = 3.75EE434 pKa = 5.22AGTSPAATPIAVTTDD449 pKa = 3.07APGTDD454 pKa = 4.02AADD457 pKa = 3.53QQHH460 pKa = 6.18FSPYY464 pKa = 10.04FEE466 pKa = 4.05MWLPSSQNLVQTVEE480 pKa = 4.4DD481 pKa = 3.67VGLTAVTLAFVLGTGPDD498 pKa = 3.45QIGWGGLGSIDD509 pKa = 5.26NDD511 pKa = 3.64TLANGTTISSMVAALQQNGVEE532 pKa = 4.33VTISFGGGYY541 pKa = 8.99GQEE544 pKa = 4.11PALSFTNVAQLTAAYY559 pKa = 9.58QSVMDD564 pKa = 4.61KK565 pKa = 11.47YY566 pKa = 11.44NVTSLDD572 pKa = 3.38FDD574 pKa = 3.92IEE576 pKa = 4.41ADD578 pKa = 3.48ALTNTAASHH587 pKa = 6.1LRR589 pKa = 11.84NEE591 pKa = 4.29ALVALEE597 pKa = 4.1QANPDD602 pKa = 3.51LTVSFTLPALPTGLNQAGLDD622 pKa = 3.72LLAQAKK628 pKa = 10.26ADD630 pKa = 3.49GVEE633 pKa = 3.87IDD635 pKa = 3.95TVNIMVMNYY644 pKa = 9.07GAYY647 pKa = 9.93YY648 pKa = 10.69DD649 pKa = 4.12SGDD652 pKa = 3.68MGKK655 pKa = 10.07DD656 pKa = 3.15AIDD659 pKa = 3.58AAEE662 pKa = 3.8ATIAQLHH669 pKa = 5.07QLGLDD674 pKa = 3.35AKK676 pKa = 10.46VAITPMIGQNDD687 pKa = 3.57VPGEE691 pKa = 4.22VFTLDD696 pKa = 4.66DD697 pKa = 3.85ARR699 pKa = 11.84QLLDD703 pKa = 3.41YY704 pKa = 11.56AEE706 pKa = 4.92GNDD709 pKa = 4.05HH710 pKa = 6.81IAYY713 pKa = 9.35IAMWSLGRR721 pKa = 11.84DD722 pKa = 3.07HH723 pKa = 7.29GDD725 pKa = 3.38DD726 pKa = 3.55VGHH729 pKa = 6.37LTEE732 pKa = 4.57TSSGVAQHH740 pKa = 7.29DD741 pKa = 3.83YY742 pKa = 11.47DD743 pKa = 3.73FAKK746 pKa = 10.12IFSMVV751 pKa = 2.75
Molecular weight: 77.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A7IGN8|IF3_XANP2 Translation initiation factor IF-3 OS=Xanthobacter autotrophicus (strain ATCC BAA-1158 / Py2) OX=78245 GN=infC PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.26IIAARR34 pKa = 11.84RR35 pKa = 11.84NHH37 pKa = 4.53GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.08
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.26IIAARR34 pKa = 11.84RR35 pKa = 11.84NHH37 pKa = 4.53GRR39 pKa = 11.84QRR41 pKa = 11.84LSAA44 pKa = 4.08
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1618488 |
35 |
3208 |
325.6 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.793 ± 0.059 | 0.803 ± 0.009 |
5.556 ± 0.028 | 5.548 ± 0.036 |
3.671 ± 0.022 | 8.761 ± 0.037 |
1.995 ± 0.016 | 4.583 ± 0.027 |
3.069 ± 0.029 | 10.319 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.015 | 2.201 ± 0.019 |
5.688 ± 0.033 | 2.881 ± 0.021 |
7.427 ± 0.04 | 5.055 ± 0.021 |
5.226 ± 0.026 | 7.847 ± 0.026 |
1.246 ± 0.012 | 2.069 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
