
Circoviridae 2 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
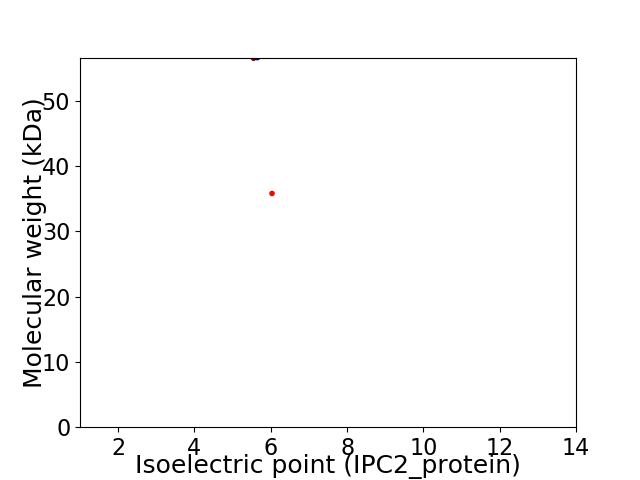
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5TNA9|S5TNA9_9CIRC ATP-dependent helicase Rep OS=Circoviridae 2 LDMD-2013 OX=1379695 PE=3 SV=1
MM1 pKa = 7.47PRR3 pKa = 11.84AKK5 pKa = 9.83RR6 pKa = 11.84VRR8 pKa = 11.84NQEE11 pKa = 3.92GEE13 pKa = 4.28VEE15 pKa = 4.3DD16 pKa = 4.34PVAKK20 pKa = 10.22GRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 10.57GMWASMKK31 pKa = 10.26LSKK34 pKa = 10.36AQPWDD39 pKa = 3.3TYY41 pKa = 9.29GARR44 pKa = 11.84RR45 pKa = 11.84VQRR48 pKa = 11.84GTPEE52 pKa = 3.8NLEE55 pKa = 4.17LFGPTWKK62 pKa = 10.4EE63 pKa = 3.02ADD65 pKa = 3.28EE66 pKa = 4.32MQRR69 pKa = 11.84SARR72 pKa = 11.84KK73 pKa = 8.54EE74 pKa = 3.6FGYY77 pKa = 10.14SGRR80 pKa = 11.84GLYY83 pKa = 10.52GPLAYY88 pKa = 10.1AAGAGIDD95 pKa = 3.72YY96 pKa = 10.56AVKK99 pKa = 10.37NPRR102 pKa = 11.84ATRR105 pKa = 11.84SFFSKK110 pKa = 10.35AFKK113 pKa = 10.53GRR115 pKa = 11.84GMYY118 pKa = 9.12TGRR121 pKa = 11.84GMYY124 pKa = 9.0TGRR127 pKa = 11.84GEE129 pKa = 4.07YY130 pKa = 8.72TASNEE135 pKa = 3.84LMAGSTSSPPMFQSAGDD152 pKa = 3.53EE153 pKa = 4.24AGALMVSHH161 pKa = 6.99RR162 pKa = 11.84EE163 pKa = 3.83YY164 pKa = 11.09VGDD167 pKa = 3.81IFAPAAADD175 pKa = 3.37VSKK178 pKa = 9.62FTVQSFPLNPGLEE191 pKa = 4.09QTFPWLSQIAQNYY204 pKa = 8.25EE205 pKa = 3.89EE206 pKa = 5.46YY207 pKa = 10.45EE208 pKa = 4.0LKK210 pKa = 10.34QCVFEE215 pKa = 4.72FVSTVQDD222 pKa = 3.24INSTNGQVGTIITATQYY239 pKa = 11.13NPSEE243 pKa = 4.18GDD245 pKa = 3.56FTDD248 pKa = 3.93KK249 pKa = 10.8PAMAAYY255 pKa = 9.24AHH257 pKa = 6.07SVSGKK262 pKa = 8.42STDD265 pKa = 3.55NQTHH269 pKa = 5.46GVEE272 pKa = 4.32CDD274 pKa = 3.17PAKK277 pKa = 10.86LSGAEE282 pKa = 3.98GKK284 pKa = 9.3YY285 pKa = 10.13VRR287 pKa = 11.84ANPVMTGEE295 pKa = 4.06DD296 pKa = 3.85LKK298 pKa = 10.38TYY300 pKa = 10.76DD301 pKa = 3.44HH302 pKa = 6.93GRR304 pKa = 11.84FQLATHH310 pKa = 7.11NIPAAMASGTLGEE323 pKa = 4.96LYY325 pKa = 10.41VAYY328 pKa = 8.31TVCLRR333 pKa = 11.84KK334 pKa = 9.65PKK336 pKa = 10.32FFVGRR341 pKa = 11.84GLGLTRR347 pKa = 11.84YY348 pKa = 9.97CEE350 pKa = 4.13VWDD353 pKa = 3.89ATASGSTTDD362 pKa = 3.49GDD364 pKa = 3.96HH365 pKa = 7.47PYY367 pKa = 10.89GADD370 pKa = 3.38GEE372 pKa = 4.53QFLGKK377 pKa = 10.06QNNIAIKK384 pKa = 8.37TVKK387 pKa = 7.93TTDD390 pKa = 2.98TLEE393 pKa = 4.17IVFPAYY399 pKa = 10.59YY400 pKa = 10.4GGNLEE405 pKa = 4.43IKK407 pKa = 9.83LANEE411 pKa = 4.29GTLSSGTFCGEE422 pKa = 4.23LVASMAVEE430 pKa = 4.53GNCVLVKK437 pKa = 10.47DD438 pKa = 5.36LYY440 pKa = 11.53ASGPSNTDD448 pKa = 2.3SPAASRR454 pKa = 11.84SAGDD458 pKa = 3.93ANNSHH463 pKa = 7.25AIIHH467 pKa = 5.46VRR469 pKa = 11.84VSPSTNAVDD478 pKa = 3.47NKK480 pKa = 10.94LIITSGQGSVFNVNQSMIDD499 pKa = 3.27VTEE502 pKa = 4.12YY503 pKa = 9.74NTYY506 pKa = 10.49GQDD509 pKa = 3.11TLEE512 pKa = 4.59LVNSAGTSVVIAA524 pKa = 4.46
MM1 pKa = 7.47PRR3 pKa = 11.84AKK5 pKa = 9.83RR6 pKa = 11.84VRR8 pKa = 11.84NQEE11 pKa = 3.92GEE13 pKa = 4.28VEE15 pKa = 4.3DD16 pKa = 4.34PVAKK20 pKa = 10.22GRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 10.57GMWASMKK31 pKa = 10.26LSKK34 pKa = 10.36AQPWDD39 pKa = 3.3TYY41 pKa = 9.29GARR44 pKa = 11.84RR45 pKa = 11.84VQRR48 pKa = 11.84GTPEE52 pKa = 3.8NLEE55 pKa = 4.17LFGPTWKK62 pKa = 10.4EE63 pKa = 3.02ADD65 pKa = 3.28EE66 pKa = 4.32MQRR69 pKa = 11.84SARR72 pKa = 11.84KK73 pKa = 8.54EE74 pKa = 3.6FGYY77 pKa = 10.14SGRR80 pKa = 11.84GLYY83 pKa = 10.52GPLAYY88 pKa = 10.1AAGAGIDD95 pKa = 3.72YY96 pKa = 10.56AVKK99 pKa = 10.37NPRR102 pKa = 11.84ATRR105 pKa = 11.84SFFSKK110 pKa = 10.35AFKK113 pKa = 10.53GRR115 pKa = 11.84GMYY118 pKa = 9.12TGRR121 pKa = 11.84GMYY124 pKa = 9.0TGRR127 pKa = 11.84GEE129 pKa = 4.07YY130 pKa = 8.72TASNEE135 pKa = 3.84LMAGSTSSPPMFQSAGDD152 pKa = 3.53EE153 pKa = 4.24AGALMVSHH161 pKa = 6.99RR162 pKa = 11.84EE163 pKa = 3.83YY164 pKa = 11.09VGDD167 pKa = 3.81IFAPAAADD175 pKa = 3.37VSKK178 pKa = 9.62FTVQSFPLNPGLEE191 pKa = 4.09QTFPWLSQIAQNYY204 pKa = 8.25EE205 pKa = 3.89EE206 pKa = 5.46YY207 pKa = 10.45EE208 pKa = 4.0LKK210 pKa = 10.34QCVFEE215 pKa = 4.72FVSTVQDD222 pKa = 3.24INSTNGQVGTIITATQYY239 pKa = 11.13NPSEE243 pKa = 4.18GDD245 pKa = 3.56FTDD248 pKa = 3.93KK249 pKa = 10.8PAMAAYY255 pKa = 9.24AHH257 pKa = 6.07SVSGKK262 pKa = 8.42STDD265 pKa = 3.55NQTHH269 pKa = 5.46GVEE272 pKa = 4.32CDD274 pKa = 3.17PAKK277 pKa = 10.86LSGAEE282 pKa = 3.98GKK284 pKa = 9.3YY285 pKa = 10.13VRR287 pKa = 11.84ANPVMTGEE295 pKa = 4.06DD296 pKa = 3.85LKK298 pKa = 10.38TYY300 pKa = 10.76DD301 pKa = 3.44HH302 pKa = 6.93GRR304 pKa = 11.84FQLATHH310 pKa = 7.11NIPAAMASGTLGEE323 pKa = 4.96LYY325 pKa = 10.41VAYY328 pKa = 8.31TVCLRR333 pKa = 11.84KK334 pKa = 9.65PKK336 pKa = 10.32FFVGRR341 pKa = 11.84GLGLTRR347 pKa = 11.84YY348 pKa = 9.97CEE350 pKa = 4.13VWDD353 pKa = 3.89ATASGSTTDD362 pKa = 3.49GDD364 pKa = 3.96HH365 pKa = 7.47PYY367 pKa = 10.89GADD370 pKa = 3.38GEE372 pKa = 4.53QFLGKK377 pKa = 10.06QNNIAIKK384 pKa = 8.37TVKK387 pKa = 7.93TTDD390 pKa = 2.98TLEE393 pKa = 4.17IVFPAYY399 pKa = 10.59YY400 pKa = 10.4GGNLEE405 pKa = 4.43IKK407 pKa = 9.83LANEE411 pKa = 4.29GTLSSGTFCGEE422 pKa = 4.23LVASMAVEE430 pKa = 4.53GNCVLVKK437 pKa = 10.47DD438 pKa = 5.36LYY440 pKa = 11.53ASGPSNTDD448 pKa = 2.3SPAASRR454 pKa = 11.84SAGDD458 pKa = 3.93ANNSHH463 pKa = 7.25AIIHH467 pKa = 5.46VRR469 pKa = 11.84VSPSTNAVDD478 pKa = 3.47NKK480 pKa = 10.94LIITSGQGSVFNVNQSMIDD499 pKa = 3.27VTEE502 pKa = 4.12YY503 pKa = 9.74NTYY506 pKa = 10.49GQDD509 pKa = 3.11TLEE512 pKa = 4.59LVNSAGTSVVIAA524 pKa = 4.46
Molecular weight: 56.47 kDa
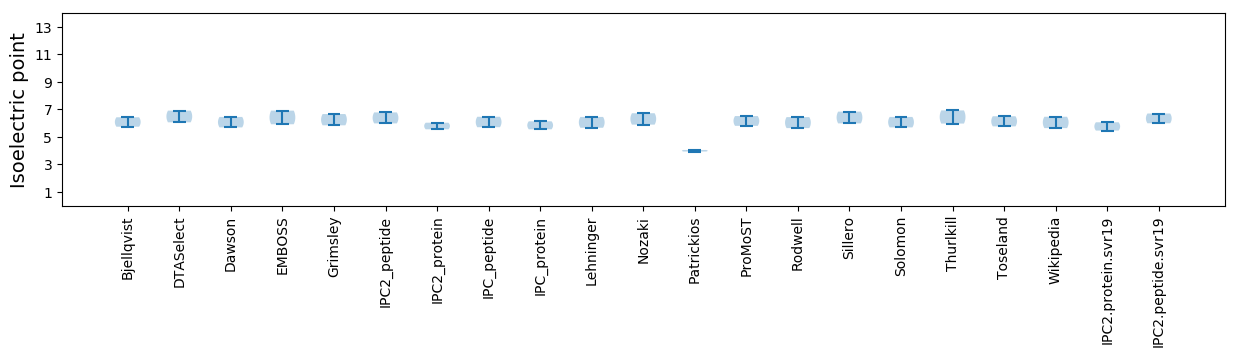
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TNA9|S5TNA9_9CIRC ATP-dependent helicase Rep OS=Circoviridae 2 LDMD-2013 OX=1379695 PE=3 SV=1
MM1 pKa = 7.83KK2 pKa = 10.21IPDD5 pKa = 3.71RR6 pKa = 11.84SRR8 pKa = 11.84HH9 pKa = 4.69RR10 pKa = 11.84AYY12 pKa = 10.9CITLNNYY19 pKa = 9.35FEE21 pKa = 5.63HH22 pKa = 7.53DD23 pKa = 3.66RR24 pKa = 11.84QNLEE28 pKa = 3.96TLVEE32 pKa = 4.08RR33 pKa = 11.84GIAKK37 pKa = 7.95YY38 pKa = 10.91VCFQPEE44 pKa = 4.23KK45 pKa = 10.95APGTGTRR52 pKa = 11.84HH53 pKa = 4.36IQGYY57 pKa = 7.56VVFQNARR64 pKa = 11.84TFSGAKK70 pKa = 10.08AIIGEE75 pKa = 4.36GAHH78 pKa = 6.28IEE80 pKa = 4.22VARR83 pKa = 11.84GSSQANIEE91 pKa = 4.2YY92 pKa = 10.21CSKK95 pKa = 11.07DD96 pKa = 3.3EE97 pKa = 5.09SRR99 pKa = 11.84DD100 pKa = 3.51TEE102 pKa = 4.08AGFGFIEE109 pKa = 4.27RR110 pKa = 11.84GNRR113 pKa = 11.84EE114 pKa = 4.17DD115 pKa = 3.84VLGTGSGGGTRR126 pKa = 11.84TDD128 pKa = 3.71LAVVAASLRR137 pKa = 11.84TGATLSQIAEE147 pKa = 4.33DD148 pKa = 4.5HH149 pKa = 5.97PASYY153 pKa = 11.01IMYY156 pKa = 8.6TRR158 pKa = 11.84GIHH161 pKa = 7.01SYY163 pKa = 10.93AQLSLPKK170 pKa = 9.46RR171 pKa = 11.84SHH173 pKa = 5.2KK174 pKa = 9.76TIVHH178 pKa = 6.56WYY180 pKa = 7.38WGPTGTGKK188 pKa = 8.08TRR190 pKa = 11.84LACEE194 pKa = 4.37EE195 pKa = 4.5SPDD198 pKa = 4.22AYY200 pKa = 9.3WKK202 pKa = 10.77SSAHH206 pKa = 4.65QWYY209 pKa = 10.03DD210 pKa = 3.28GYY212 pKa = 11.46DD213 pKa = 3.4GLADD217 pKa = 5.13IIIDD221 pKa = 4.05DD222 pKa = 4.15YY223 pKa = 11.45RR224 pKa = 11.84CSFSTFNEE232 pKa = 3.88LLRR235 pKa = 11.84LLDD238 pKa = 4.26RR239 pKa = 11.84YY240 pKa = 9.93PYY242 pKa = 9.02QAQVKK247 pKa = 9.95GGTIHH252 pKa = 6.74VNAKK256 pKa = 10.08RR257 pKa = 11.84IFITAPKK264 pKa = 9.43DD265 pKa = 3.5PRR267 pKa = 11.84SMWQSRR273 pKa = 11.84TEE275 pKa = 3.64EE276 pKa = 5.05DD277 pKa = 2.87IAQLEE282 pKa = 4.08RR283 pKa = 11.84RR284 pKa = 11.84IEE286 pKa = 3.92VVRR289 pKa = 11.84YY290 pKa = 8.92FGVDD294 pKa = 3.25EE295 pKa = 5.26NIPEE299 pKa = 4.31PVLNEE304 pKa = 3.86APAVIHH310 pKa = 6.57GFDD313 pKa = 3.93PGVALLL319 pKa = 4.31
MM1 pKa = 7.83KK2 pKa = 10.21IPDD5 pKa = 3.71RR6 pKa = 11.84SRR8 pKa = 11.84HH9 pKa = 4.69RR10 pKa = 11.84AYY12 pKa = 10.9CITLNNYY19 pKa = 9.35FEE21 pKa = 5.63HH22 pKa = 7.53DD23 pKa = 3.66RR24 pKa = 11.84QNLEE28 pKa = 3.96TLVEE32 pKa = 4.08RR33 pKa = 11.84GIAKK37 pKa = 7.95YY38 pKa = 10.91VCFQPEE44 pKa = 4.23KK45 pKa = 10.95APGTGTRR52 pKa = 11.84HH53 pKa = 4.36IQGYY57 pKa = 7.56VVFQNARR64 pKa = 11.84TFSGAKK70 pKa = 10.08AIIGEE75 pKa = 4.36GAHH78 pKa = 6.28IEE80 pKa = 4.22VARR83 pKa = 11.84GSSQANIEE91 pKa = 4.2YY92 pKa = 10.21CSKK95 pKa = 11.07DD96 pKa = 3.3EE97 pKa = 5.09SRR99 pKa = 11.84DD100 pKa = 3.51TEE102 pKa = 4.08AGFGFIEE109 pKa = 4.27RR110 pKa = 11.84GNRR113 pKa = 11.84EE114 pKa = 4.17DD115 pKa = 3.84VLGTGSGGGTRR126 pKa = 11.84TDD128 pKa = 3.71LAVVAASLRR137 pKa = 11.84TGATLSQIAEE147 pKa = 4.33DD148 pKa = 4.5HH149 pKa = 5.97PASYY153 pKa = 11.01IMYY156 pKa = 8.6TRR158 pKa = 11.84GIHH161 pKa = 7.01SYY163 pKa = 10.93AQLSLPKK170 pKa = 9.46RR171 pKa = 11.84SHH173 pKa = 5.2KK174 pKa = 9.76TIVHH178 pKa = 6.56WYY180 pKa = 7.38WGPTGTGKK188 pKa = 8.08TRR190 pKa = 11.84LACEE194 pKa = 4.37EE195 pKa = 4.5SPDD198 pKa = 4.22AYY200 pKa = 9.3WKK202 pKa = 10.77SSAHH206 pKa = 4.65QWYY209 pKa = 10.03DD210 pKa = 3.28GYY212 pKa = 11.46DD213 pKa = 3.4GLADD217 pKa = 5.13IIIDD221 pKa = 4.05DD222 pKa = 4.15YY223 pKa = 11.45RR224 pKa = 11.84CSFSTFNEE232 pKa = 3.88LLRR235 pKa = 11.84LLDD238 pKa = 4.26RR239 pKa = 11.84YY240 pKa = 9.93PYY242 pKa = 9.02QAQVKK247 pKa = 9.95GGTIHH252 pKa = 6.74VNAKK256 pKa = 10.08RR257 pKa = 11.84IFITAPKK264 pKa = 9.43DD265 pKa = 3.5PRR267 pKa = 11.84SMWQSRR273 pKa = 11.84TEE275 pKa = 3.64EE276 pKa = 5.05DD277 pKa = 2.87IAQLEE282 pKa = 4.08RR283 pKa = 11.84RR284 pKa = 11.84IEE286 pKa = 3.92VVRR289 pKa = 11.84YY290 pKa = 8.92FGVDD294 pKa = 3.25EE295 pKa = 5.26NIPEE299 pKa = 4.31PVLNEE304 pKa = 3.86APAVIHH310 pKa = 6.57GFDD313 pKa = 3.93PGVALLL319 pKa = 4.31
Molecular weight: 35.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
843 |
319 |
524 |
421.5 |
46.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.727 ± 0.55 | 1.305 ± 0.152 |
5.101 ± 0.314 | 6.287 ± 0.353 |
3.677 ± 0.133 | 9.727 ± 0.55 |
2.254 ± 0.692 | 4.745 ± 1.429 |
4.389 ± 0.364 | 5.813 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.017 ± 0.624 | 4.27 ± 0.658 |
4.508 ± 0.069 | 3.796 ± 0.02 |
5.813 ± 1.173 | 7.236 ± 0.56 |
7.117 ± 0.491 | 6.287 ± 0.555 |
1.186 ± 0.221 | 4.745 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
