
Acinetobacter brisouii CIP 110357
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter; Acinetobacter brisouii
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
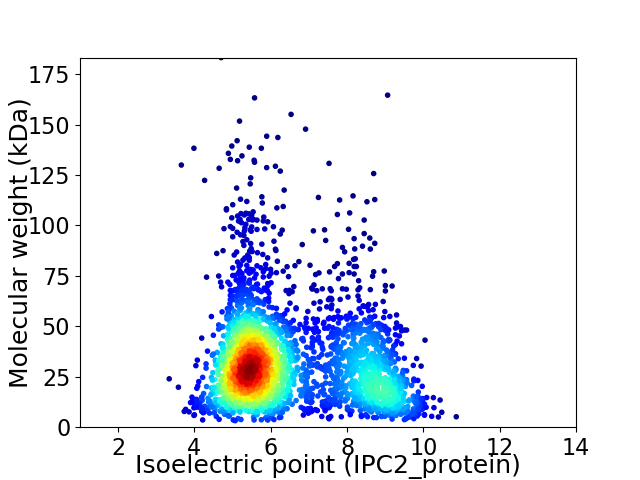
Virtual 2D-PAGE plot for 2927 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V2UKC3|V2UKC3_9GAMM Thioredoxin domain-containing protein OS=Acinetobacter brisouii CIP 110357 OX=1341683 GN=P255_02785 PE=4 SV=1
YYY2 pKa = 10.09IDDD5 pKa = 3.76IQFSDDD11 pKa = 3.65TTLDDD16 pKa = 3.0EEE18 pKa = 4.35VKKK21 pKa = 10.81LVLLGTEEE29 pKa = 4.07EE30 pKa = 4.81DDD32 pKa = 3.42LRR34 pKa = 11.84AYYY37 pKa = 7.05TGSTISGEEE46 pKa = 4.02GDDD49 pKa = 5.61DD50 pKa = 3.89IHHH53 pKa = 6.68NIGSDDD59 pKa = 3.37IYYY62 pKa = 10.67GDDD65 pKa = 4.18QDDD68 pKa = 4.14LYYY71 pKa = 10.37DDD73 pKa = 4.59GNDDD77 pKa = 3.6HH78 pKa = 7.14EEE80 pKa = 4.43GADDD84 pKa = 3.58DDD86 pKa = 3.53LYYY89 pKa = 10.74GNGNDDD95 pKa = 3.52LIGGSGNDDD104 pKa = 4.32LDDD107 pKa = 4.64GDDD110 pKa = 5.53SDDD113 pKa = 3.46LYYY116 pKa = 10.83GTGDDD121 pKa = 4.79DD122 pKa = 4.15LNGGAGNDDD131 pKa = 4.01LDDD134 pKa = 4.96GDDD137 pKa = 5.43SDDD140 pKa = 3.82LNGGIGDDD148 pKa = 5.04DD149 pKa = 3.64LVGGTGDDD157 pKa = 3.68DD158 pKa = 3.77LIGGQGNDDD167 pKa = 3.85YYY169 pKa = 11.18FDDD172 pKa = 5.74GWGQDDD178 pKa = 3.58IQYYY182 pKa = 8.96EE183 pKa = 4.8KKK185 pKa = 10.38QTDDD189 pKa = 3.02DDD191 pKa = 4.07IEEE194 pKa = 5.16ININPDDD201 pKa = 3.33DD202 pKa = 5.11IVRR205 pKa = 11.84KKK207 pKa = 9.9GQDDD211 pKa = 3.06VVMHHH216 pKa = 7.58ITGDDD221 pKa = 3.27LIVEEE226 pKa = 4.82QFSDDD231 pKa = 3.88YY232 pKa = 11.34SNKKK236 pKa = 9.06INKKK240 pKa = 8.54RR241 pKa = 11.84FDDD244 pKa = 4.92DD245 pKa = 3.66TEEE248 pKa = 3.93DDD250 pKa = 3.74EE251 pKa = 5.06DD252 pKa = 3.87LNVQAVKKK260 pKa = 9.33TEEE263 pKa = 3.82DDD265 pKa = 4.5DD266 pKa = 4.96IEEE269 pKa = 4.5TTDDD273 pKa = 3.02HH274 pKa = 7.24DD275 pKa = 4.87IHHH278 pKa = 7.08GAGDDD283 pKa = 4.2DD284 pKa = 3.89IYYY287 pKa = 9.92RR288 pKa = 11.84PVYYY292 pKa = 10.85EEE294 pKa = 4.02PEEE297 pKa = 3.9QYYY300 pKa = 11.19YY301 pKa = 9.74YYY303 pKa = 10.74GEEE306 pKa = 4.45NDDD309 pKa = 3.57KK310 pKa = 10.43YYY312 pKa = 10.21TGYYY316 pKa = 10.28DDD318 pKa = 4.73GVGDDD323 pKa = 5.42DD324 pKa = 4.12EE325 pKa = 5.5EEE327 pKa = 4.54SGHHH331 pKa = 6.78LGGEEE336 pKa = 4.32NDDD339 pKa = 3.23LSGQGVLEEE348 pKa = 4.41QSGNDDD354 pKa = 2.9LTGQGKKK361 pKa = 10.37YYY363 pKa = 10.96GDDD366 pKa = 3.66QDDD369 pKa = 3.22LTLTGLANDDD379 pKa = 3.98EE380 pKa = 4.76GLLSGGAGDDD390 pKa = 5.21DD391 pKa = 3.66LTIDDD396 pKa = 3.98NQMDDD401 pKa = 4.72VNEEE405 pKa = 3.84YY406 pKa = 10.54QDDD409 pKa = 2.96FYYY412 pKa = 11.08DDD414 pKa = 2.86DDD416 pKa = 4.09YYY418 pKa = 11.4HH419 pKa = 7.31IQDDD423 pKa = 3.41EEE425 pKa = 4.06TEEE428 pKa = 4.06QRR430 pKa = 11.84AVYYY434 pKa = 10.64EEE436 pKa = 4.62GQGNDDD442 pKa = 3.31IYYY445 pKa = 11.02SFGDDD450 pKa = 4.01EE451 pKa = 3.66YYY453 pKa = 10.96FNLGDDD459 pKa = 3.93KKK461 pKa = 9.88DD462 pKa = 3.82IIEEE466 pKa = 4.08PAGQNYYY473 pKa = 10.37NVAVSFDDD481 pKa = 3.55LRR483 pKa = 11.84FGQGITTIDDD493 pKa = 3.38SLHHH497 pKa = 6.34YYY499 pKa = 7.24TDDD502 pKa = 3.25VIKKK506 pKa = 8.73HH507 pKa = 6.03NQTDDD512 pKa = 3.26ITIQNYYY519 pKa = 7.09TDDD522 pKa = 3.41HHH524 pKa = 6.35HH525 pKa = 6.55KK526 pKa = 10.54NEEE529 pKa = 3.91QFADDD534 pKa = 3.49TIWNNAYYY542 pKa = 9.63EEE544 pKa = 4.06HHH546 pKa = 5.01TYYY549 pKa = 10.53HH550 pKa = 5.82TANNDDD556 pKa = 3.53EE557 pKa = 4.13WGYYY561 pKa = 11.35NTDDD565 pKa = 3.05EE566 pKa = 4.47FEEE569 pKa = 4.23DDD571 pKa = 3.7GDDD574 pKa = 3.85DD575 pKa = 3.7KK576 pKa = 11.71YYY578 pKa = 11.0GAGNDDD584 pKa = 3.83IYYY587 pKa = 9.75QAGEEE592 pKa = 4.75DD593 pKa = 3.8LWGQAGNDDD602 pKa = 3.63LYYY605 pKa = 11.24GIGADDD611 pKa = 3.18YY612 pKa = 11.34EEE614 pKa = 5.01NEEE617 pKa = 4.94DDD619 pKa = 4.56DD620 pKa = 4.85LYYY623 pKa = 10.73DDD625 pKa = 4.04DDD627 pKa = 3.41DDD629 pKa = 3.2LYYY632 pKa = 10.55GAGNDDD638 pKa = 3.2LYYY641 pKa = 10.42GQGDDD646 pKa = 4.52DD647 pKa = 3.12LRR649 pKa = 11.84GGTCHHH655 pKa = 6.9DD656 pKa = 4.36LDDD659 pKa = 4.12GAGNDDD665 pKa = 3.12YYY667 pKa = 11.47YYY669 pKa = 11.2YY670 pKa = 10.68ADDD673 pKa = 3.59TEEE676 pKa = 4.76IDDD679 pKa = 3.38TGGGTDDD686 pKa = 3.74LWLMDDD692 pKa = 3.49GITEEE697 pKa = 4.39DD698 pKa = 3.78IKKK701 pKa = 10.0TKKK704 pKa = 10.13EE705 pKa = 3.39NDDD708 pKa = 4.24IVTIDDD714 pKa = 3.6NPNQSVRR721 pKa = 11.84VKKK724 pKa = 10.81DD725 pKa = 3.55HH726 pKa = 6.43LGGEEE731 pKa = 3.97KK732 pKa = 10.07ISSVQPNGGYYY743 pKa = 9.12ITAAQIAVKKK753 pKa = 10.46NASAGGLSDDD763 pKa = 4.31EEE765 pKa = 5.64DDD767 pKa = 3.75IYYY770 pKa = 10.78YYY772 pKa = 8.18TGAMTITEEE781 pKa = 3.96SGNDDD786 pKa = 3.02VLFKKK791 pKa = 11.37GITFSQVGNYYY802 pKa = 7.2TKKK805 pKa = 10.89GDDD808 pKa = 3.74DD809 pKa = 4.56ILKKK813 pKa = 10.0NGSNTNKKK821 pKa = 8.44TIKKK825 pKa = 10.89FFLAGNYYY833 pKa = 9.38VEEE836 pKa = 4.23LQFEEE841 pKa = 4.55GGQLTAEEE849 pKa = 4.37IFGAFGLTIPSGTTPIPPNPVGDDD873 pKa = 3.69TYYY876 pKa = 10.57YYY878 pKa = 11.79SGEEE882 pKa = 3.9TITEEE887 pKa = 3.75SGNDDD892 pKa = 3.11KK893 pKa = 11.23IFKKK897 pKa = 10.95GITFNQVGNYYY908 pKa = 6.74TKKK911 pKa = 10.89GDDD914 pKa = 3.74DD915 pKa = 4.56ILKKK919 pKa = 10.08NGSDDD924 pKa = 3.22NTVIVKKK931 pKa = 10.0FFLAGQYYY939 pKa = 10.14VEEE942 pKa = 4.28FEEE945 pKa = 4.38EEE947 pKa = 3.96GGKKK951 pKa = 7.63TAEEE955 pKa = 4.06IFGAFGITMPQQSAPANTSPEEE977 pKa = 3.99MDDD980 pKa = 4.28DDD982 pKa = 4.27SNTIYYY988 pKa = 10.66YYY990 pKa = 10.66SGSMIIDDD998 pKa = 3.39EE999 pKa = 4.69LGTDDD1004 pKa = 3.13VVFGNGISFSQVGNYYY1020 pKa = 7.45TQSGDDD1026 pKa = 3.73DD1027 pKa = 4.87ILKKK1031 pKa = 10.05NGSNSNKKK1039 pKa = 8.24TVKKK1043 pKa = 10.9DD1044 pKa = 4.06FLGGAHHH1051 pKa = 5.56EE1052 pKa = 4.3EEE1054 pKa = 4.48FNFEEE1059 pKa = 4.06GGSISSQQIYYY1070 pKa = 8.4VFGVDDD1076 pKa = 3.37HH1077 pKa = 6.6TNAEEE1082 pKa = 4.12DD1083 pKa = 3.71EE1084 pKa = 4.58TSIVVGDDD1092 pKa = 3.89DDD1094 pKa = 6.12DD1095 pKa = 5.24ILSSNAAVSEEE1106 pKa = 4.02FVLNEEE1112 pKa = 4.04NDDD1115 pKa = 3.51LEEE1118 pKa = 4.55LLNASGEEE1126 pKa = 4.36AVDDD1130 pKa = 3.62YY1131 pKa = 10.4TDDD1134 pKa = 4.16DDD1136 pKa = 4.03TEEE1139 pKa = 4.18DD1140 pKa = 4.04IDDD1143 pKa = 4.06SQILDDD1149 pKa = 3.74QATSSNLSDDD1159 pKa = 4.25YY1160 pKa = 11.36EEE1162 pKa = 3.82TYYY1165 pKa = 9.87ANAKKK1170 pKa = 9.29NTLSLRR1176 pKa = 11.84DDD1178 pKa = 3.76LNVTVKKK1185 pKa = 10.84DD1186 pKa = 3.61LIFTNQAEEE1195 pKa = 4.36LSITDDD1201 pKa = 3.68MMNQAIIY
YYY2 pKa = 10.09IDDD5 pKa = 3.76IQFSDDD11 pKa = 3.65TTLDDD16 pKa = 3.0EEE18 pKa = 4.35VKKK21 pKa = 10.81LVLLGTEEE29 pKa = 4.07EE30 pKa = 4.81DDD32 pKa = 3.42LRR34 pKa = 11.84AYYY37 pKa = 7.05TGSTISGEEE46 pKa = 4.02GDDD49 pKa = 5.61DD50 pKa = 3.89IHHH53 pKa = 6.68NIGSDDD59 pKa = 3.37IYYY62 pKa = 10.67GDDD65 pKa = 4.18QDDD68 pKa = 4.14LYYY71 pKa = 10.37DDD73 pKa = 4.59GNDDD77 pKa = 3.6HH78 pKa = 7.14EEE80 pKa = 4.43GADDD84 pKa = 3.58DDD86 pKa = 3.53LYYY89 pKa = 10.74GNGNDDD95 pKa = 3.52LIGGSGNDDD104 pKa = 4.32LDDD107 pKa = 4.64GDDD110 pKa = 5.53SDDD113 pKa = 3.46LYYY116 pKa = 10.83GTGDDD121 pKa = 4.79DD122 pKa = 4.15LNGGAGNDDD131 pKa = 4.01LDDD134 pKa = 4.96GDDD137 pKa = 5.43SDDD140 pKa = 3.82LNGGIGDDD148 pKa = 5.04DD149 pKa = 3.64LVGGTGDDD157 pKa = 3.68DD158 pKa = 3.77LIGGQGNDDD167 pKa = 3.85YYY169 pKa = 11.18FDDD172 pKa = 5.74GWGQDDD178 pKa = 3.58IQYYY182 pKa = 8.96EE183 pKa = 4.8KKK185 pKa = 10.38QTDDD189 pKa = 3.02DDD191 pKa = 4.07IEEE194 pKa = 5.16ININPDDD201 pKa = 3.33DD202 pKa = 5.11IVRR205 pKa = 11.84KKK207 pKa = 9.9GQDDD211 pKa = 3.06VVMHHH216 pKa = 7.58ITGDDD221 pKa = 3.27LIVEEE226 pKa = 4.82QFSDDD231 pKa = 3.88YY232 pKa = 11.34SNKKK236 pKa = 9.06INKKK240 pKa = 8.54RR241 pKa = 11.84FDDD244 pKa = 4.92DD245 pKa = 3.66TEEE248 pKa = 3.93DDD250 pKa = 3.74EE251 pKa = 5.06DD252 pKa = 3.87LNVQAVKKK260 pKa = 9.33TEEE263 pKa = 3.82DDD265 pKa = 4.5DD266 pKa = 4.96IEEE269 pKa = 4.5TTDDD273 pKa = 3.02HH274 pKa = 7.24DD275 pKa = 4.87IHHH278 pKa = 7.08GAGDDD283 pKa = 4.2DD284 pKa = 3.89IYYY287 pKa = 9.92RR288 pKa = 11.84PVYYY292 pKa = 10.85EEE294 pKa = 4.02PEEE297 pKa = 3.9QYYY300 pKa = 11.19YY301 pKa = 9.74YYY303 pKa = 10.74GEEE306 pKa = 4.45NDDD309 pKa = 3.57KK310 pKa = 10.43YYY312 pKa = 10.21TGYYY316 pKa = 10.28DDD318 pKa = 4.73GVGDDD323 pKa = 5.42DD324 pKa = 4.12EE325 pKa = 5.5EEE327 pKa = 4.54SGHHH331 pKa = 6.78LGGEEE336 pKa = 4.32NDDD339 pKa = 3.23LSGQGVLEEE348 pKa = 4.41QSGNDDD354 pKa = 2.9LTGQGKKK361 pKa = 10.37YYY363 pKa = 10.96GDDD366 pKa = 3.66QDDD369 pKa = 3.22LTLTGLANDDD379 pKa = 3.98EE380 pKa = 4.76GLLSGGAGDDD390 pKa = 5.21DD391 pKa = 3.66LTIDDD396 pKa = 3.98NQMDDD401 pKa = 4.72VNEEE405 pKa = 3.84YY406 pKa = 10.54QDDD409 pKa = 2.96FYYY412 pKa = 11.08DDD414 pKa = 2.86DDD416 pKa = 4.09YYY418 pKa = 11.4HH419 pKa = 7.31IQDDD423 pKa = 3.41EEE425 pKa = 4.06TEEE428 pKa = 4.06QRR430 pKa = 11.84AVYYY434 pKa = 10.64EEE436 pKa = 4.62GQGNDDD442 pKa = 3.31IYYY445 pKa = 11.02SFGDDD450 pKa = 4.01EE451 pKa = 3.66YYY453 pKa = 10.96FNLGDDD459 pKa = 3.93KKK461 pKa = 9.88DD462 pKa = 3.82IIEEE466 pKa = 4.08PAGQNYYY473 pKa = 10.37NVAVSFDDD481 pKa = 3.55LRR483 pKa = 11.84FGQGITTIDDD493 pKa = 3.38SLHHH497 pKa = 6.34YYY499 pKa = 7.24TDDD502 pKa = 3.25VIKKK506 pKa = 8.73HH507 pKa = 6.03NQTDDD512 pKa = 3.26ITIQNYYY519 pKa = 7.09TDDD522 pKa = 3.41HHH524 pKa = 6.35HH525 pKa = 6.55KK526 pKa = 10.54NEEE529 pKa = 3.91QFADDD534 pKa = 3.49TIWNNAYYY542 pKa = 9.63EEE544 pKa = 4.06HHH546 pKa = 5.01TYYY549 pKa = 10.53HH550 pKa = 5.82TANNDDD556 pKa = 3.53EE557 pKa = 4.13WGYYY561 pKa = 11.35NTDDD565 pKa = 3.05EE566 pKa = 4.47FEEE569 pKa = 4.23DDD571 pKa = 3.7GDDD574 pKa = 3.85DD575 pKa = 3.7KK576 pKa = 11.71YYY578 pKa = 11.0GAGNDDD584 pKa = 3.83IYYY587 pKa = 9.75QAGEEE592 pKa = 4.75DD593 pKa = 3.8LWGQAGNDDD602 pKa = 3.63LYYY605 pKa = 11.24GIGADDD611 pKa = 3.18YY612 pKa = 11.34EEE614 pKa = 5.01NEEE617 pKa = 4.94DDD619 pKa = 4.56DD620 pKa = 4.85LYYY623 pKa = 10.73DDD625 pKa = 4.04DDD627 pKa = 3.41DDD629 pKa = 3.2LYYY632 pKa = 10.55GAGNDDD638 pKa = 3.2LYYY641 pKa = 10.42GQGDDD646 pKa = 4.52DD647 pKa = 3.12LRR649 pKa = 11.84GGTCHHH655 pKa = 6.9DD656 pKa = 4.36LDDD659 pKa = 4.12GAGNDDD665 pKa = 3.12YYY667 pKa = 11.47YYY669 pKa = 11.2YY670 pKa = 10.68ADDD673 pKa = 3.59TEEE676 pKa = 4.76IDDD679 pKa = 3.38TGGGTDDD686 pKa = 3.74LWLMDDD692 pKa = 3.49GITEEE697 pKa = 4.39DD698 pKa = 3.78IKKK701 pKa = 10.0TKKK704 pKa = 10.13EE705 pKa = 3.39NDDD708 pKa = 4.24IVTIDDD714 pKa = 3.6NPNQSVRR721 pKa = 11.84VKKK724 pKa = 10.81DD725 pKa = 3.55HH726 pKa = 6.43LGGEEE731 pKa = 3.97KK732 pKa = 10.07ISSVQPNGGYYY743 pKa = 9.12ITAAQIAVKKK753 pKa = 10.46NASAGGLSDDD763 pKa = 4.31EEE765 pKa = 5.64DDD767 pKa = 3.75IYYY770 pKa = 10.78YYY772 pKa = 8.18TGAMTITEEE781 pKa = 3.96SGNDDD786 pKa = 3.02VLFKKK791 pKa = 11.37GITFSQVGNYYY802 pKa = 7.2TKKK805 pKa = 10.89GDDD808 pKa = 3.74DD809 pKa = 4.56ILKKK813 pKa = 10.0NGSNTNKKK821 pKa = 8.44TIKKK825 pKa = 10.89FFLAGNYYY833 pKa = 9.38VEEE836 pKa = 4.23LQFEEE841 pKa = 4.55GGQLTAEEE849 pKa = 4.37IFGAFGLTIPSGTTPIPPNPVGDDD873 pKa = 3.69TYYY876 pKa = 10.57YYY878 pKa = 11.79SGEEE882 pKa = 3.9TITEEE887 pKa = 3.75SGNDDD892 pKa = 3.11KK893 pKa = 11.23IFKKK897 pKa = 10.95GITFNQVGNYYY908 pKa = 6.74TKKK911 pKa = 10.89GDDD914 pKa = 3.74DD915 pKa = 4.56ILKKK919 pKa = 10.08NGSDDD924 pKa = 3.22NTVIVKKK931 pKa = 10.0FFLAGQYYY939 pKa = 10.14VEEE942 pKa = 4.28FEEE945 pKa = 4.38EEE947 pKa = 3.96GGKKK951 pKa = 7.63TAEEE955 pKa = 4.06IFGAFGITMPQQSAPANTSPEEE977 pKa = 3.99MDDD980 pKa = 4.28DDD982 pKa = 4.27SNTIYYY988 pKa = 10.66YYY990 pKa = 10.66SGSMIIDDD998 pKa = 3.39EE999 pKa = 4.69LGTDDD1004 pKa = 3.13VVFGNGISFSQVGNYYY1020 pKa = 7.45TQSGDDD1026 pKa = 3.73DD1027 pKa = 4.87ILKKK1031 pKa = 10.05NGSNSNKKK1039 pKa = 8.24TVKKK1043 pKa = 10.9DD1044 pKa = 4.06FLGGAHHH1051 pKa = 5.56EE1052 pKa = 4.3EEE1054 pKa = 4.48FNFEEE1059 pKa = 4.06GGSISSQQIYYY1070 pKa = 8.4VFGVDDD1076 pKa = 3.37HH1077 pKa = 6.6TNAEEE1082 pKa = 4.12DD1083 pKa = 3.71EE1084 pKa = 4.58TSIVVGDDD1092 pKa = 3.89DDD1094 pKa = 6.12DD1095 pKa = 5.24ILSSNAAVSEEE1106 pKa = 4.02FVLNEEE1112 pKa = 4.04NDDD1115 pKa = 3.51LEEE1118 pKa = 4.55LLNASGEEE1126 pKa = 4.36AVDDD1130 pKa = 3.62YY1131 pKa = 10.4TDDD1134 pKa = 4.16DDD1136 pKa = 4.03TEEE1139 pKa = 4.18DD1140 pKa = 4.04IDDD1143 pKa = 4.06SQILDDD1149 pKa = 3.74QATSSNLSDDD1159 pKa = 4.25YY1160 pKa = 11.36EEE1162 pKa = 3.82TYYY1165 pKa = 9.87ANAKKK1170 pKa = 9.29NTLSLRR1176 pKa = 11.84DDD1178 pKa = 3.76LNVTVKKK1185 pKa = 10.84DD1186 pKa = 3.61LIFTNQAEEE1195 pKa = 4.36LSITDDD1201 pKa = 3.68MMNQAIIY
Molecular weight: 129.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V2UQ88|V2UQ88_9GAMM Uncharacterized protein OS=Acinetobacter brisouii CIP 110357 OX=1341683 GN=P255_01293 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
MM1 pKa = 7.45KK2 pKa = 9.55RR3 pKa = 11.84TFQPSEE9 pKa = 3.97LKK11 pKa = 10.13RR12 pKa = 11.84KK13 pKa = 8.98RR14 pKa = 11.84VHH16 pKa = 6.36GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4AGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.18GRR39 pKa = 11.84HH40 pKa = 5.1SLTVV44 pKa = 3.06
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896350 |
30 |
1625 |
306.2 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.925 ± 0.048 | 1.011 ± 0.015 |
5.033 ± 0.039 | 5.626 ± 0.043 |
4.17 ± 0.034 | 6.487 ± 0.051 |
2.618 ± 0.025 | 6.688 ± 0.038 |
5.159 ± 0.038 | 10.692 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.513 ± 0.023 | 4.157 ± 0.035 |
3.981 ± 0.027 | 6.056 ± 0.068 |
4.445 ± 0.035 | 5.974 ± 0.038 |
5.253 ± 0.031 | 6.746 ± 0.039 |
1.285 ± 0.02 | 3.181 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
