
Haloactinospora alba
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Haloactinospora
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
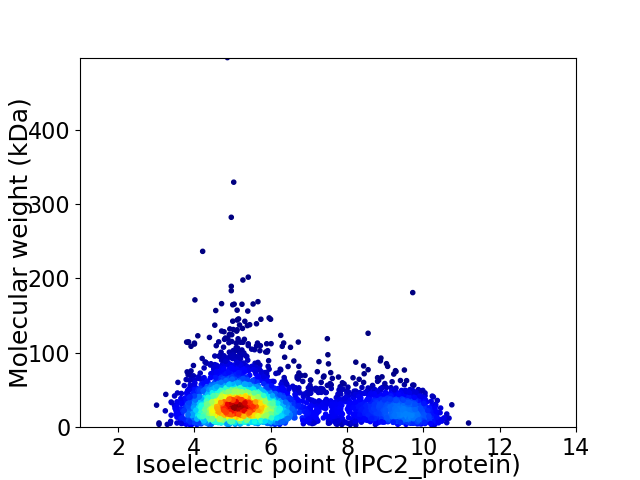
Virtual 2D-PAGE plot for 4398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A543N7F1|A0A543N7F1_9ACTN ABC-2 type transport system permease protein OS=Haloactinospora alba OX=405555 GN=FHX37_4462 PE=3 SV=1
MM1 pKa = 7.84PAPAHH6 pKa = 5.4PRR8 pKa = 11.84GWLASIAVATAAVTTAALLHH28 pKa = 6.04TPAHH32 pKa = 6.62AEE34 pKa = 4.19QNSSPDD40 pKa = 3.71TQDD43 pKa = 3.26VPSGDD48 pKa = 2.74SWQPDD53 pKa = 3.52EE54 pKa = 5.23EE55 pKa = 4.4VDD57 pKa = 3.56RR58 pKa = 11.84DD59 pKa = 3.82PSGGEE64 pKa = 3.78PPEE67 pKa = 4.45EE68 pKa = 4.43DD69 pKa = 2.34WKK71 pKa = 11.11QPHH74 pKa = 6.0EE75 pKa = 4.47RR76 pKa = 11.84EE77 pKa = 4.14ADD79 pKa = 3.63QQDD82 pKa = 3.93GLSAQDD88 pKa = 3.4TDD90 pKa = 3.86IDD92 pKa = 5.08AEE94 pKa = 4.33DD95 pKa = 3.9SCDD98 pKa = 3.53DD99 pKa = 3.46GTNRR103 pKa = 11.84GVQDD107 pKa = 3.67HH108 pKa = 6.64YY109 pKa = 11.06PLQRR113 pKa = 11.84HH114 pKa = 5.33QISDD118 pKa = 3.65RR119 pKa = 11.84LEE121 pKa = 4.92LDD123 pKa = 3.55VNLEE127 pKa = 3.9NGNTVLRR134 pKa = 11.84HH135 pKa = 6.19RR136 pKa = 11.84EE137 pKa = 3.78LTIPGTGIDD146 pKa = 3.95LSLSSVYY153 pKa = 10.57NSQDD157 pKa = 2.99LGTNGWKK164 pKa = 10.69LNTGTDD170 pKa = 3.16VGLDD174 pKa = 4.36FITDD178 pKa = 3.29SDD180 pKa = 4.01DD181 pKa = 3.42VVFRR185 pKa = 11.84GPSGFCEE192 pKa = 4.24TFTDD196 pKa = 3.94QGDD199 pKa = 3.85GTFASPEE206 pKa = 3.95DD207 pKa = 3.67MEE209 pKa = 7.23ADD211 pKa = 4.97LEE213 pKa = 4.16EE214 pKa = 5.44LDD216 pKa = 3.5NGRR219 pKa = 11.84YY220 pKa = 9.84ALTFHH225 pKa = 6.39QGPYY229 pKa = 9.74ADD231 pKa = 4.02QMWTFTADD239 pKa = 2.53GWLYY243 pKa = 10.89AQSDD247 pKa = 4.14HH248 pKa = 6.83NGNTTTLNYY257 pKa = 10.64DD258 pKa = 3.52DD259 pKa = 6.09DD260 pKa = 4.81GLITSITDD268 pKa = 3.18DD269 pKa = 3.31QDD271 pKa = 3.44RR272 pKa = 11.84VTEE275 pKa = 4.45LDD277 pKa = 2.8WHH279 pKa = 5.34QNRR282 pKa = 11.84IGLTEE287 pKa = 3.83ITDD290 pKa = 3.78PTGATAADD298 pKa = 3.67YY299 pKa = 10.91SYY301 pKa = 11.63NADD304 pKa = 3.57GQLTEE309 pKa = 3.88ITDD312 pKa = 3.62RR313 pKa = 11.84AGNDD317 pKa = 2.65ITFAYY322 pKa = 10.25DD323 pKa = 3.58DD324 pKa = 4.37NNHH327 pKa = 6.36LSEE330 pKa = 3.98ITNARR335 pKa = 11.84GHH337 pKa = 5.57TWNLDD342 pKa = 3.01HH343 pKa = 7.51DD344 pKa = 4.42TAGQLTEE351 pKa = 4.74LDD353 pKa = 3.65QPVTGDD359 pKa = 3.11EE360 pKa = 4.33RR361 pKa = 11.84AVTTYY366 pKa = 11.44DD367 pKa = 2.9HH368 pKa = 7.07GEE370 pKa = 4.17AEE372 pKa = 4.59TEE374 pKa = 4.2VTDD377 pKa = 4.16PRR379 pKa = 11.84GNTSTHH385 pKa = 5.8TFDD388 pKa = 3.17EE389 pKa = 4.29QGRR392 pKa = 11.84QEE394 pKa = 4.28EE395 pKa = 4.43ATDD398 pKa = 3.76PEE400 pKa = 5.06GNTRR404 pKa = 11.84STTWTAAASVAAATDD419 pKa = 3.6PAGASTTYY427 pKa = 10.79DD428 pKa = 3.53YY429 pKa = 11.85DD430 pKa = 3.98DD431 pKa = 4.98LNNMVGTQLPSGAEE445 pKa = 3.97YY446 pKa = 10.66QIGYY450 pKa = 10.07ADD452 pKa = 3.43SANPHH457 pKa = 6.56KK458 pKa = 9.12PTSITAPEE466 pKa = 4.17EE467 pKa = 4.11EE468 pKa = 4.63VEE470 pKa = 4.84LDD472 pKa = 3.26YY473 pKa = 11.53DD474 pKa = 4.21DD475 pKa = 5.92RR476 pKa = 11.84GNLTQIRR483 pKa = 11.84RR484 pKa = 11.84PGAEE488 pKa = 3.95EE489 pKa = 4.18PEE491 pKa = 4.1QSLEE495 pKa = 3.95YY496 pKa = 10.89NNNGTLASQTNGQGAKK512 pKa = 9.39TEE514 pKa = 4.05FSYY517 pKa = 11.58DD518 pKa = 3.19EE519 pKa = 5.23DD520 pKa = 4.77GNLTEE525 pKa = 5.51IDD527 pKa = 3.69QPDD530 pKa = 3.94PLGTTSFTYY539 pKa = 10.41DD540 pKa = 2.85ALSRR544 pKa = 11.84VTSVTDD550 pKa = 3.76GNGTTLEE557 pKa = 4.05YY558 pKa = 10.38AYY560 pKa = 10.67DD561 pKa = 3.57KK562 pKa = 10.8LDD564 pKa = 3.45RR565 pKa = 11.84VVEE568 pKa = 4.24IVHH571 pKa = 6.98DD572 pKa = 3.96GDD574 pKa = 3.48IRR576 pKa = 11.84QSTEE580 pKa = 3.49YY581 pKa = 10.57NPNGDD586 pKa = 3.11VTATHH591 pKa = 6.97TPQATVAHH599 pKa = 7.47AYY601 pKa = 9.39NKK603 pKa = 10.12RR604 pKa = 11.84GEE606 pKa = 4.25VANTGRR612 pKa = 11.84DD613 pKa = 3.36DD614 pKa = 4.37GQDD617 pKa = 3.45HH618 pKa = 6.93EE619 pKa = 5.58SYY621 pKa = 10.96DD622 pKa = 3.63YY623 pKa = 11.14TYY625 pKa = 11.23DD626 pKa = 3.25QAGNLTSLSEE636 pKa = 4.23HH637 pKa = 6.8GATTDD642 pKa = 3.65YY643 pKa = 11.28AYY645 pKa = 9.19DD646 pKa = 3.46TANRR650 pKa = 11.84LTTVTDD656 pKa = 3.77DD657 pKa = 3.79SGGEE661 pKa = 3.92TTIGYY666 pKa = 10.04DD667 pKa = 3.11EE668 pKa = 4.95AGNRR672 pKa = 11.84DD673 pKa = 4.59EE674 pKa = 4.9ITFPDD679 pKa = 4.64GATQSLDD686 pKa = 3.4HH687 pKa = 6.85NEE689 pKa = 4.11AGLLTEE695 pKa = 4.85SVMANADD702 pKa = 3.39GDD704 pKa = 4.37TLAEE708 pKa = 3.93AAYY711 pKa = 10.4SYY713 pKa = 11.21TDD715 pKa = 5.2DD716 pKa = 5.79DD717 pKa = 4.68GQDD720 pKa = 3.31TTKK723 pKa = 10.79LQSRR727 pKa = 11.84TVNGQSRR734 pKa = 11.84EE735 pKa = 3.87FTYY738 pKa = 10.57DD739 pKa = 2.64EE740 pKa = 4.38RR741 pKa = 11.84GRR743 pKa = 11.84LTSDD747 pKa = 2.33GHH749 pKa = 5.72TDD751 pKa = 3.43YY752 pKa = 11.14TYY754 pKa = 11.45DD755 pKa = 3.55EE756 pKa = 4.88ADD758 pKa = 3.57NLTSAGDD765 pKa = 3.15TDD767 pKa = 4.15YY768 pKa = 11.44QVNKK772 pKa = 9.69ADD774 pKa = 3.65QVTEE778 pKa = 3.89VDD780 pKa = 3.97DD781 pKa = 4.11TEE783 pKa = 5.27LGYY786 pKa = 10.86DD787 pKa = 3.24QAGNLTEE794 pKa = 5.55AGQTDD799 pKa = 3.73IDD801 pKa = 4.2YY802 pKa = 11.0SPTGQMVDD810 pKa = 3.23KK811 pKa = 10.85DD812 pKa = 4.1AGNSASDD819 pKa = 3.64LQVSYY824 pKa = 8.96DD825 pKa = 3.83TADD828 pKa = 3.1STQRR832 pKa = 11.84RR833 pKa = 11.84TITQGSGDD841 pKa = 3.96NQTEE845 pKa = 4.1HH846 pKa = 6.65TLTNTALGISSIEE859 pKa = 3.81QDD861 pKa = 3.3DD862 pKa = 4.18GDD864 pKa = 3.58RR865 pKa = 11.84TRR867 pKa = 11.84YY868 pKa = 9.57VRR870 pKa = 11.84DD871 pKa = 3.05PDD873 pKa = 3.51GRR875 pKa = 11.84MLGMITGGQRR885 pKa = 11.84YY886 pKa = 8.11NAATDD891 pKa = 3.82AQNSTLALAEE901 pKa = 5.24DD902 pKa = 4.61GTEE905 pKa = 4.09STDD908 pKa = 3.26PDD910 pKa = 3.33VAYY913 pKa = 10.32DD914 pKa = 3.59YY915 pKa = 10.89TPYY918 pKa = 10.95GQTEE922 pKa = 4.16TATAGSGDD930 pKa = 3.49QAAQTNPFTYY940 pKa = 9.73TGAYY944 pKa = 9.58EE945 pKa = 4.62LDD947 pKa = 3.94NGDD950 pKa = 3.3KK951 pKa = 11.03ALGHH955 pKa = 6.92RR956 pKa = 11.84YY957 pKa = 9.65LSQRR961 pKa = 11.84THH963 pKa = 7.24RR964 pKa = 11.84FTQQDD969 pKa = 3.79PSWQEE974 pKa = 3.76DD975 pKa = 3.4NLYY978 pKa = 10.13TYY980 pKa = 10.99AEE982 pKa = 4.6CDD984 pKa = 4.49PINKK988 pKa = 8.51MDD990 pKa = 4.27RR991 pKa = 11.84NGLSACGWAIGDD1003 pKa = 5.02FITGYY1008 pKa = 10.01ISYY1011 pKa = 9.63HH1012 pKa = 5.04FAVYY1016 pKa = 9.82GVAALLAGTVAVAAAPIAIAAVSTGIAIYY1045 pKa = 10.78GMGRR1049 pKa = 11.84GIAGMYY1055 pKa = 9.42HH1056 pKa = 6.01SCC1058 pKa = 4.77
MM1 pKa = 7.84PAPAHH6 pKa = 5.4PRR8 pKa = 11.84GWLASIAVATAAVTTAALLHH28 pKa = 6.04TPAHH32 pKa = 6.62AEE34 pKa = 4.19QNSSPDD40 pKa = 3.71TQDD43 pKa = 3.26VPSGDD48 pKa = 2.74SWQPDD53 pKa = 3.52EE54 pKa = 5.23EE55 pKa = 4.4VDD57 pKa = 3.56RR58 pKa = 11.84DD59 pKa = 3.82PSGGEE64 pKa = 3.78PPEE67 pKa = 4.45EE68 pKa = 4.43DD69 pKa = 2.34WKK71 pKa = 11.11QPHH74 pKa = 6.0EE75 pKa = 4.47RR76 pKa = 11.84EE77 pKa = 4.14ADD79 pKa = 3.63QQDD82 pKa = 3.93GLSAQDD88 pKa = 3.4TDD90 pKa = 3.86IDD92 pKa = 5.08AEE94 pKa = 4.33DD95 pKa = 3.9SCDD98 pKa = 3.53DD99 pKa = 3.46GTNRR103 pKa = 11.84GVQDD107 pKa = 3.67HH108 pKa = 6.64YY109 pKa = 11.06PLQRR113 pKa = 11.84HH114 pKa = 5.33QISDD118 pKa = 3.65RR119 pKa = 11.84LEE121 pKa = 4.92LDD123 pKa = 3.55VNLEE127 pKa = 3.9NGNTVLRR134 pKa = 11.84HH135 pKa = 6.19RR136 pKa = 11.84EE137 pKa = 3.78LTIPGTGIDD146 pKa = 3.95LSLSSVYY153 pKa = 10.57NSQDD157 pKa = 2.99LGTNGWKK164 pKa = 10.69LNTGTDD170 pKa = 3.16VGLDD174 pKa = 4.36FITDD178 pKa = 3.29SDD180 pKa = 4.01DD181 pKa = 3.42VVFRR185 pKa = 11.84GPSGFCEE192 pKa = 4.24TFTDD196 pKa = 3.94QGDD199 pKa = 3.85GTFASPEE206 pKa = 3.95DD207 pKa = 3.67MEE209 pKa = 7.23ADD211 pKa = 4.97LEE213 pKa = 4.16EE214 pKa = 5.44LDD216 pKa = 3.5NGRR219 pKa = 11.84YY220 pKa = 9.84ALTFHH225 pKa = 6.39QGPYY229 pKa = 9.74ADD231 pKa = 4.02QMWTFTADD239 pKa = 2.53GWLYY243 pKa = 10.89AQSDD247 pKa = 4.14HH248 pKa = 6.83NGNTTTLNYY257 pKa = 10.64DD258 pKa = 3.52DD259 pKa = 6.09DD260 pKa = 4.81GLITSITDD268 pKa = 3.18DD269 pKa = 3.31QDD271 pKa = 3.44RR272 pKa = 11.84VTEE275 pKa = 4.45LDD277 pKa = 2.8WHH279 pKa = 5.34QNRR282 pKa = 11.84IGLTEE287 pKa = 3.83ITDD290 pKa = 3.78PTGATAADD298 pKa = 3.67YY299 pKa = 10.91SYY301 pKa = 11.63NADD304 pKa = 3.57GQLTEE309 pKa = 3.88ITDD312 pKa = 3.62RR313 pKa = 11.84AGNDD317 pKa = 2.65ITFAYY322 pKa = 10.25DD323 pKa = 3.58DD324 pKa = 4.37NNHH327 pKa = 6.36LSEE330 pKa = 3.98ITNARR335 pKa = 11.84GHH337 pKa = 5.57TWNLDD342 pKa = 3.01HH343 pKa = 7.51DD344 pKa = 4.42TAGQLTEE351 pKa = 4.74LDD353 pKa = 3.65QPVTGDD359 pKa = 3.11EE360 pKa = 4.33RR361 pKa = 11.84AVTTYY366 pKa = 11.44DD367 pKa = 2.9HH368 pKa = 7.07GEE370 pKa = 4.17AEE372 pKa = 4.59TEE374 pKa = 4.2VTDD377 pKa = 4.16PRR379 pKa = 11.84GNTSTHH385 pKa = 5.8TFDD388 pKa = 3.17EE389 pKa = 4.29QGRR392 pKa = 11.84QEE394 pKa = 4.28EE395 pKa = 4.43ATDD398 pKa = 3.76PEE400 pKa = 5.06GNTRR404 pKa = 11.84STTWTAAASVAAATDD419 pKa = 3.6PAGASTTYY427 pKa = 10.79DD428 pKa = 3.53YY429 pKa = 11.85DD430 pKa = 3.98DD431 pKa = 4.98LNNMVGTQLPSGAEE445 pKa = 3.97YY446 pKa = 10.66QIGYY450 pKa = 10.07ADD452 pKa = 3.43SANPHH457 pKa = 6.56KK458 pKa = 9.12PTSITAPEE466 pKa = 4.17EE467 pKa = 4.11EE468 pKa = 4.63VEE470 pKa = 4.84LDD472 pKa = 3.26YY473 pKa = 11.53DD474 pKa = 4.21DD475 pKa = 5.92RR476 pKa = 11.84GNLTQIRR483 pKa = 11.84RR484 pKa = 11.84PGAEE488 pKa = 3.95EE489 pKa = 4.18PEE491 pKa = 4.1QSLEE495 pKa = 3.95YY496 pKa = 10.89NNNGTLASQTNGQGAKK512 pKa = 9.39TEE514 pKa = 4.05FSYY517 pKa = 11.58DD518 pKa = 3.19EE519 pKa = 5.23DD520 pKa = 4.77GNLTEE525 pKa = 5.51IDD527 pKa = 3.69QPDD530 pKa = 3.94PLGTTSFTYY539 pKa = 10.41DD540 pKa = 2.85ALSRR544 pKa = 11.84VTSVTDD550 pKa = 3.76GNGTTLEE557 pKa = 4.05YY558 pKa = 10.38AYY560 pKa = 10.67DD561 pKa = 3.57KK562 pKa = 10.8LDD564 pKa = 3.45RR565 pKa = 11.84VVEE568 pKa = 4.24IVHH571 pKa = 6.98DD572 pKa = 3.96GDD574 pKa = 3.48IRR576 pKa = 11.84QSTEE580 pKa = 3.49YY581 pKa = 10.57NPNGDD586 pKa = 3.11VTATHH591 pKa = 6.97TPQATVAHH599 pKa = 7.47AYY601 pKa = 9.39NKK603 pKa = 10.12RR604 pKa = 11.84GEE606 pKa = 4.25VANTGRR612 pKa = 11.84DD613 pKa = 3.36DD614 pKa = 4.37GQDD617 pKa = 3.45HH618 pKa = 6.93EE619 pKa = 5.58SYY621 pKa = 10.96DD622 pKa = 3.63YY623 pKa = 11.14TYY625 pKa = 11.23DD626 pKa = 3.25QAGNLTSLSEE636 pKa = 4.23HH637 pKa = 6.8GATTDD642 pKa = 3.65YY643 pKa = 11.28AYY645 pKa = 9.19DD646 pKa = 3.46TANRR650 pKa = 11.84LTTVTDD656 pKa = 3.77DD657 pKa = 3.79SGGEE661 pKa = 3.92TTIGYY666 pKa = 10.04DD667 pKa = 3.11EE668 pKa = 4.95AGNRR672 pKa = 11.84DD673 pKa = 4.59EE674 pKa = 4.9ITFPDD679 pKa = 4.64GATQSLDD686 pKa = 3.4HH687 pKa = 6.85NEE689 pKa = 4.11AGLLTEE695 pKa = 4.85SVMANADD702 pKa = 3.39GDD704 pKa = 4.37TLAEE708 pKa = 3.93AAYY711 pKa = 10.4SYY713 pKa = 11.21TDD715 pKa = 5.2DD716 pKa = 5.79DD717 pKa = 4.68GQDD720 pKa = 3.31TTKK723 pKa = 10.79LQSRR727 pKa = 11.84TVNGQSRR734 pKa = 11.84EE735 pKa = 3.87FTYY738 pKa = 10.57DD739 pKa = 2.64EE740 pKa = 4.38RR741 pKa = 11.84GRR743 pKa = 11.84LTSDD747 pKa = 2.33GHH749 pKa = 5.72TDD751 pKa = 3.43YY752 pKa = 11.14TYY754 pKa = 11.45DD755 pKa = 3.55EE756 pKa = 4.88ADD758 pKa = 3.57NLTSAGDD765 pKa = 3.15TDD767 pKa = 4.15YY768 pKa = 11.44QVNKK772 pKa = 9.69ADD774 pKa = 3.65QVTEE778 pKa = 3.89VDD780 pKa = 3.97DD781 pKa = 4.11TEE783 pKa = 5.27LGYY786 pKa = 10.86DD787 pKa = 3.24QAGNLTEE794 pKa = 5.55AGQTDD799 pKa = 3.73IDD801 pKa = 4.2YY802 pKa = 11.0SPTGQMVDD810 pKa = 3.23KK811 pKa = 10.85DD812 pKa = 4.1AGNSASDD819 pKa = 3.64LQVSYY824 pKa = 8.96DD825 pKa = 3.83TADD828 pKa = 3.1STQRR832 pKa = 11.84RR833 pKa = 11.84TITQGSGDD841 pKa = 3.96NQTEE845 pKa = 4.1HH846 pKa = 6.65TLTNTALGISSIEE859 pKa = 3.81QDD861 pKa = 3.3DD862 pKa = 4.18GDD864 pKa = 3.58RR865 pKa = 11.84TRR867 pKa = 11.84YY868 pKa = 9.57VRR870 pKa = 11.84DD871 pKa = 3.05PDD873 pKa = 3.51GRR875 pKa = 11.84MLGMITGGQRR885 pKa = 11.84YY886 pKa = 8.11NAATDD891 pKa = 3.82AQNSTLALAEE901 pKa = 5.24DD902 pKa = 4.61GTEE905 pKa = 4.09STDD908 pKa = 3.26PDD910 pKa = 3.33VAYY913 pKa = 10.32DD914 pKa = 3.59YY915 pKa = 10.89TPYY918 pKa = 10.95GQTEE922 pKa = 4.16TATAGSGDD930 pKa = 3.49QAAQTNPFTYY940 pKa = 9.73TGAYY944 pKa = 9.58EE945 pKa = 4.62LDD947 pKa = 3.94NGDD950 pKa = 3.3KK951 pKa = 11.03ALGHH955 pKa = 6.92RR956 pKa = 11.84YY957 pKa = 9.65LSQRR961 pKa = 11.84THH963 pKa = 7.24RR964 pKa = 11.84FTQQDD969 pKa = 3.79PSWQEE974 pKa = 3.76DD975 pKa = 3.4NLYY978 pKa = 10.13TYY980 pKa = 10.99AEE982 pKa = 4.6CDD984 pKa = 4.49PINKK988 pKa = 8.51MDD990 pKa = 4.27RR991 pKa = 11.84NGLSACGWAIGDD1003 pKa = 5.02FITGYY1008 pKa = 10.01ISYY1011 pKa = 9.63HH1012 pKa = 5.04FAVYY1016 pKa = 9.82GVAALLAGTVAVAAAPIAIAAVSTGIAIYY1045 pKa = 10.78GMGRR1049 pKa = 11.84GIAGMYY1055 pKa = 9.42HH1056 pKa = 6.01SCC1058 pKa = 4.77
Molecular weight: 114.69 kDa
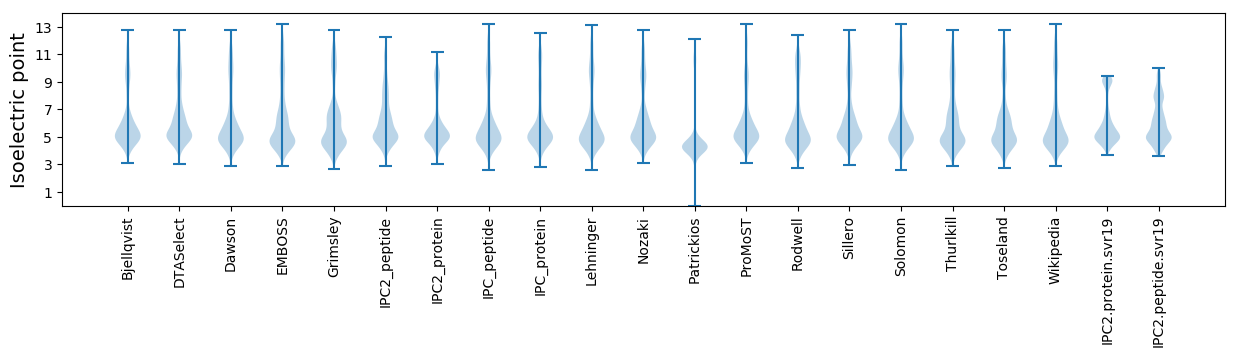
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A543N9X2|A0A543N9X2_9ACTN Uncharacterized protein OS=Haloactinospora alba OX=405555 GN=FHX37_3949 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.5GFRR20 pKa = 11.84KK21 pKa = 9.9RR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIISARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.3GRR40 pKa = 11.84ARR42 pKa = 11.84LTVSHH47 pKa = 7.05
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.5GFRR20 pKa = 11.84KK21 pKa = 9.9RR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIISARR35 pKa = 11.84RR36 pKa = 11.84NKK38 pKa = 9.3GRR40 pKa = 11.84ARR42 pKa = 11.84LTVSHH47 pKa = 7.05
Molecular weight: 5.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1363651 |
30 |
4669 |
310.1 |
33.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.402 ± 0.059 | 0.805 ± 0.011 |
6.116 ± 0.04 | 6.65 ± 0.037 |
2.688 ± 0.024 | 9.152 ± 0.034 |
2.407 ± 0.019 | 3.124 ± 0.026 |
1.645 ± 0.025 | 9.918 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.887 ± 0.015 | 2.042 ± 0.022 |
5.944 ± 0.03 | 2.937 ± 0.025 |
8.168 ± 0.04 | 5.825 ± 0.027 |
6.27 ± 0.028 | 8.469 ± 0.041 |
1.5 ± 0.017 | 2.051 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
