
Aeromicrobium sp. PE09-221
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; unclassified Aeromicrobium
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
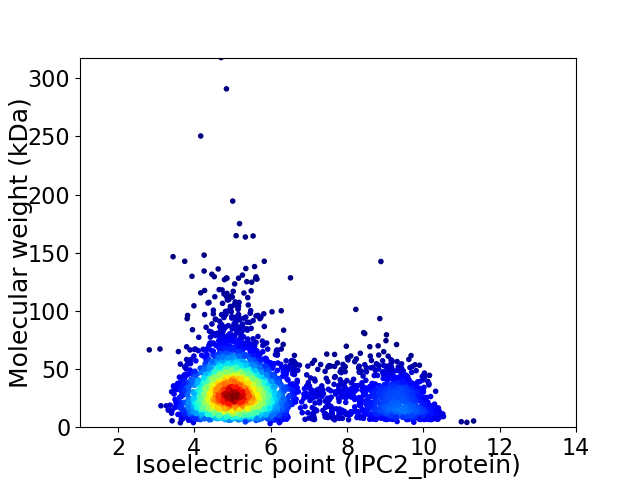
Virtual 2D-PAGE plot for 3780 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A200H9M9|A0A200H9M9_9ACTN Anti-sigma factor antagonist OS=Aeromicrobium sp. PE09-221 OX=1898043 GN=BHE97_10925 PE=3 SV=1
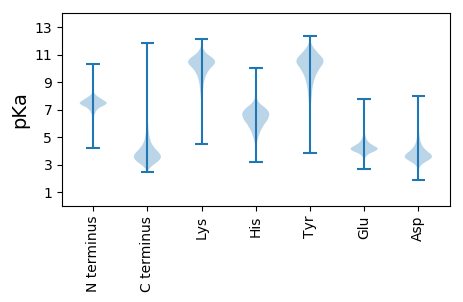
MM1 pKa = 7.12NRR3 pKa = 11.84SAAIVLGAAGIVLALIVAAAVYY25 pKa = 10.35VIGDD29 pKa = 3.56WDD31 pKa = 4.2GDD33 pKa = 4.02DD34 pKa = 4.3PAQSATPTGEE44 pKa = 3.96QAPEE48 pKa = 4.07GLDD51 pKa = 3.17AFYY54 pKa = 10.73DD55 pKa = 3.74QQIDD59 pKa = 3.67WEE61 pKa = 4.62SCDD64 pKa = 4.74SGDD67 pKa = 4.45ARR69 pKa = 11.84CATAEE74 pKa = 4.17VPVDD78 pKa = 3.51YY79 pKa = 10.36DD80 pKa = 3.83DD81 pKa = 5.51PSGEE85 pKa = 4.37TIDD88 pKa = 4.61LALLKK93 pKa = 10.78VEE95 pKa = 4.48ATGEE99 pKa = 4.03RR100 pKa = 11.84QGTIFVNPGGPGGSAQDD117 pKa = 3.55FALMMGARR125 pKa = 11.84FPSSVRR131 pKa = 11.84EE132 pKa = 4.06AYY134 pKa = 10.3DD135 pKa = 3.19IVGMDD140 pKa = 3.48PRR142 pKa = 11.84GVGDD146 pKa = 3.98STPLQCLDD154 pKa = 4.62DD155 pKa = 5.02ADD157 pKa = 4.06FDD159 pKa = 4.69AFIALDD165 pKa = 4.36PDD167 pKa = 4.05PEE169 pKa = 4.37TPEE172 pKa = 4.25EE173 pKa = 4.13IEE175 pKa = 3.99EE176 pKa = 4.09SRR178 pKa = 11.84EE179 pKa = 3.97GIEE182 pKa = 5.7AMGEE186 pKa = 3.97ACRR189 pKa = 11.84EE190 pKa = 4.01NSGDD194 pKa = 3.61LAAHH198 pKa = 6.42VSTAEE203 pKa = 3.89VARR206 pKa = 11.84DD207 pKa = 3.45HH208 pKa = 7.15DD209 pKa = 4.35VVRR212 pKa = 11.84AALGEE217 pKa = 4.18EE218 pKa = 4.07TFNWFGASYY227 pKa = 8.11GTQLGAQYY235 pKa = 11.64AEE237 pKa = 4.2MFPDD241 pKa = 3.01RR242 pKa = 11.84VGRR245 pKa = 11.84MVLDD249 pKa = 4.38GGLDD253 pKa = 3.54PALSSIEE260 pKa = 4.0VSLGQAEE267 pKa = 4.73GFQRR271 pKa = 11.84ALDD274 pKa = 4.21AYY276 pKa = 9.84IADD279 pKa = 4.38CVSSGDD285 pKa = 3.98CPLGSSEE292 pKa = 6.05DD293 pKa = 3.76EE294 pKa = 4.49ALDD297 pKa = 3.67AVAGFMADD305 pKa = 4.25LEE307 pKa = 4.29QEE309 pKa = 4.28PLVSRR314 pKa = 11.84GGRR317 pKa = 11.84EE318 pKa = 3.96LTEE321 pKa = 3.89GLAFYY326 pKa = 10.55GVAVTLYY333 pKa = 10.7AKK335 pKa = 10.15EE336 pKa = 3.91SWSYY340 pKa = 9.45LTAAFEE346 pKa = 4.52AALSDD351 pKa = 4.16DD352 pKa = 4.2DD353 pKa = 5.22PGVFLQLADD362 pKa = 3.37IYY364 pKa = 10.91FDD366 pKa = 3.61RR367 pKa = 11.84NARR370 pKa = 11.84GEE372 pKa = 4.05FQDD375 pKa = 4.02NSGQAIYY382 pKa = 10.49AVNCLDD388 pKa = 3.94GPLDD392 pKa = 3.59VTVEE396 pKa = 4.21EE397 pKa = 5.43VEE399 pKa = 4.89SEE401 pKa = 3.67WLPRR405 pKa = 11.84FRR407 pKa = 11.84EE408 pKa = 4.02ASPVFGGALGWGVMACADD426 pKa = 3.75WPIEE430 pKa = 4.09STNPQEE436 pKa = 3.96PVAAEE441 pKa = 3.98GADD444 pKa = 3.78PIVVVGTTRR453 pKa = 11.84DD454 pKa = 3.22PATPYY459 pKa = 10.18EE460 pKa = 4.31WSQSLAEE467 pKa = 4.21EE468 pKa = 4.47LSSGVLVTRR477 pKa = 11.84EE478 pKa = 4.13GDD480 pKa = 3.19GHH482 pKa = 6.18TGYY485 pKa = 11.73GMGNACVDD493 pKa = 3.77EE494 pKa = 5.39AINAYY499 pKa = 9.79LVEE502 pKa = 4.29GTVPDD507 pKa = 4.98DD508 pKa = 4.14GLVCAEE514 pKa = 4.17
MM1 pKa = 7.12NRR3 pKa = 11.84SAAIVLGAAGIVLALIVAAAVYY25 pKa = 10.35VIGDD29 pKa = 3.56WDD31 pKa = 4.2GDD33 pKa = 4.02DD34 pKa = 4.3PAQSATPTGEE44 pKa = 3.96QAPEE48 pKa = 4.07GLDD51 pKa = 3.17AFYY54 pKa = 10.73DD55 pKa = 3.74QQIDD59 pKa = 3.67WEE61 pKa = 4.62SCDD64 pKa = 4.74SGDD67 pKa = 4.45ARR69 pKa = 11.84CATAEE74 pKa = 4.17VPVDD78 pKa = 3.51YY79 pKa = 10.36DD80 pKa = 3.83DD81 pKa = 5.51PSGEE85 pKa = 4.37TIDD88 pKa = 4.61LALLKK93 pKa = 10.78VEE95 pKa = 4.48ATGEE99 pKa = 4.03RR100 pKa = 11.84QGTIFVNPGGPGGSAQDD117 pKa = 3.55FALMMGARR125 pKa = 11.84FPSSVRR131 pKa = 11.84EE132 pKa = 4.06AYY134 pKa = 10.3DD135 pKa = 3.19IVGMDD140 pKa = 3.48PRR142 pKa = 11.84GVGDD146 pKa = 3.98STPLQCLDD154 pKa = 4.62DD155 pKa = 5.02ADD157 pKa = 4.06FDD159 pKa = 4.69AFIALDD165 pKa = 4.36PDD167 pKa = 4.05PEE169 pKa = 4.37TPEE172 pKa = 4.25EE173 pKa = 4.13IEE175 pKa = 3.99EE176 pKa = 4.09SRR178 pKa = 11.84EE179 pKa = 3.97GIEE182 pKa = 5.7AMGEE186 pKa = 3.97ACRR189 pKa = 11.84EE190 pKa = 4.01NSGDD194 pKa = 3.61LAAHH198 pKa = 6.42VSTAEE203 pKa = 3.89VARR206 pKa = 11.84DD207 pKa = 3.45HH208 pKa = 7.15DD209 pKa = 4.35VVRR212 pKa = 11.84AALGEE217 pKa = 4.18EE218 pKa = 4.07TFNWFGASYY227 pKa = 8.11GTQLGAQYY235 pKa = 11.64AEE237 pKa = 4.2MFPDD241 pKa = 3.01RR242 pKa = 11.84VGRR245 pKa = 11.84MVLDD249 pKa = 4.38GGLDD253 pKa = 3.54PALSSIEE260 pKa = 4.0VSLGQAEE267 pKa = 4.73GFQRR271 pKa = 11.84ALDD274 pKa = 4.21AYY276 pKa = 9.84IADD279 pKa = 4.38CVSSGDD285 pKa = 3.98CPLGSSEE292 pKa = 6.05DD293 pKa = 3.76EE294 pKa = 4.49ALDD297 pKa = 3.67AVAGFMADD305 pKa = 4.25LEE307 pKa = 4.29QEE309 pKa = 4.28PLVSRR314 pKa = 11.84GGRR317 pKa = 11.84EE318 pKa = 3.96LTEE321 pKa = 3.89GLAFYY326 pKa = 10.55GVAVTLYY333 pKa = 10.7AKK335 pKa = 10.15EE336 pKa = 3.91SWSYY340 pKa = 9.45LTAAFEE346 pKa = 4.52AALSDD351 pKa = 4.16DD352 pKa = 4.2DD353 pKa = 5.22PGVFLQLADD362 pKa = 3.37IYY364 pKa = 10.91FDD366 pKa = 3.61RR367 pKa = 11.84NARR370 pKa = 11.84GEE372 pKa = 4.05FQDD375 pKa = 4.02NSGQAIYY382 pKa = 10.49AVNCLDD388 pKa = 3.94GPLDD392 pKa = 3.59VTVEE396 pKa = 4.21EE397 pKa = 5.43VEE399 pKa = 4.89SEE401 pKa = 3.67WLPRR405 pKa = 11.84FRR407 pKa = 11.84EE408 pKa = 4.02ASPVFGGALGWGVMACADD426 pKa = 3.75WPIEE430 pKa = 4.09STNPQEE436 pKa = 3.96PVAAEE441 pKa = 3.98GADD444 pKa = 3.78PIVVVGTTRR453 pKa = 11.84DD454 pKa = 3.22PATPYY459 pKa = 10.18EE460 pKa = 4.31WSQSLAEE467 pKa = 4.21EE468 pKa = 4.47LSSGVLVTRR477 pKa = 11.84EE478 pKa = 4.13GDD480 pKa = 3.19GHH482 pKa = 6.18TGYY485 pKa = 11.73GMGNACVDD493 pKa = 3.77EE494 pKa = 5.39AINAYY499 pKa = 9.79LVEE502 pKa = 4.29GTVPDD507 pKa = 4.98DD508 pKa = 4.14GLVCAEE514 pKa = 4.17
Molecular weight: 54.25 kDa
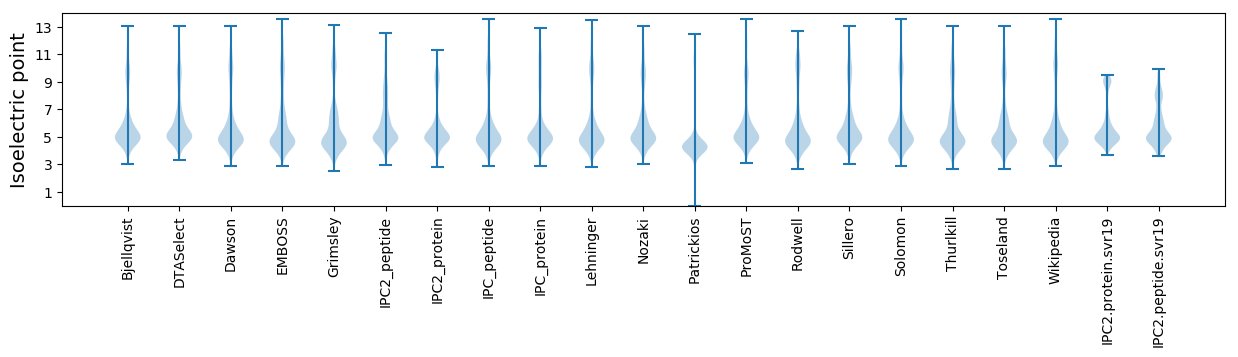
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A200HK11|A0A200HK11_9ACTN ABC transmembrane type-1 domain-containing protein OS=Aeromicrobium sp. PE09-221 OX=1898043 GN=BHE97_00320 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.97KK16 pKa = 9.33HH17 pKa = 4.25GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.74GRR40 pKa = 11.84ANLSAA45 pKa = 4.66
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1179454 |
29 |
3016 |
312.0 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.627 ± 0.056 | 0.659 ± 0.01 |
6.49 ± 0.034 | 6.278 ± 0.041 |
2.928 ± 0.022 | 8.969 ± 0.037 |
2.169 ± 0.02 | 4.456 ± 0.027 |
1.813 ± 0.03 | 10.146 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.942 ± 0.017 | 1.784 ± 0.022 |
5.282 ± 0.025 | 2.822 ± 0.022 |
7.677 ± 0.047 | 5.534 ± 0.028 |
6.015 ± 0.029 | 8.988 ± 0.037 |
1.493 ± 0.017 | 1.926 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
