
Eidolon helvum papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Psipapillomavirus; Psipapillomavirus 3
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
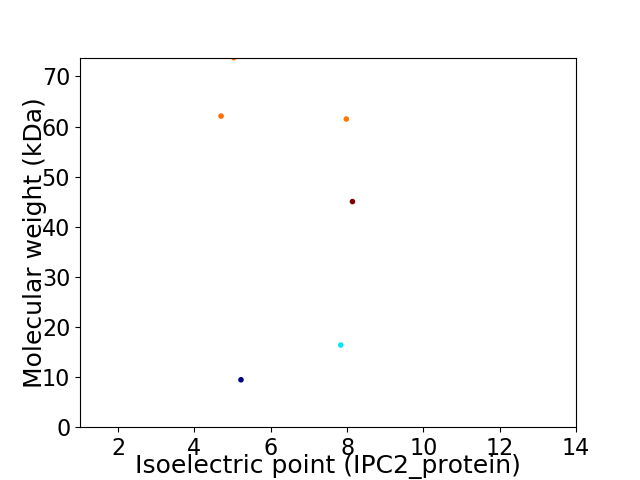
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P8YVV7|A0A1P8YVV7_9PAPI Regulatory protein E2 OS=Eidolon helvum papillomavirus 3 OX=1335477 GN=E2 PE=3 SV=1
MM1 pKa = 7.41HH2 pKa = 6.97PAKK5 pKa = 10.15RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VPRR11 pKa = 11.84DD12 pKa = 3.01SAEE15 pKa = 3.93NIYY18 pKa = 10.51RR19 pKa = 11.84NCIHH23 pKa = 7.18FGTCPPDD30 pKa = 3.41VVNKK34 pKa = 10.4IEE36 pKa = 4.33GKK38 pKa = 8.15TVADD42 pKa = 6.06KK43 pKa = 10.81ILQWGSAGVFLGNLGISTGKK63 pKa = 10.75GSANPLGYY71 pKa = 10.54VPLGRR76 pKa = 11.84GGGVGVGAWPRR87 pKa = 11.84PGLATRR93 pKa = 11.84PVVPTAPIEE102 pKa = 4.09AVGPIDD108 pKa = 3.64VGPAVVDD115 pKa = 3.68PAVVDD120 pKa = 3.71PVLIEE125 pKa = 4.23MEE127 pKa = 5.37PILPRR132 pKa = 11.84HH133 pKa = 6.12PEE135 pKa = 3.94VVDD138 pKa = 4.48EE139 pKa = 4.74IVPIAPSDD147 pKa = 3.58PSVISDD153 pKa = 3.23NGVYY157 pKa = 9.31PAHH160 pKa = 7.05PAVIDD165 pKa = 3.84EE166 pKa = 4.43SLPTGGGRR174 pKa = 11.84TVEE177 pKa = 4.44VVAEE181 pKa = 4.07VHH183 pKa = 6.67HH184 pKa = 6.49PADD187 pKa = 4.14YY188 pKa = 10.79FPVHH192 pKa = 6.93PIDD195 pKa = 3.96TGGSDD200 pKa = 3.61NPAIIDD206 pKa = 3.63VGEE209 pKa = 5.33DD210 pKa = 3.02IPLLPRR216 pKa = 11.84TSTPHH221 pKa = 5.65HH222 pKa = 6.79TDD224 pKa = 2.94PAILHH229 pKa = 5.99ATTSFGTSAEE239 pKa = 4.27VGGARR244 pKa = 11.84YY245 pKa = 7.61TVVDD249 pKa = 5.46FEE251 pKa = 4.4ATDD254 pKa = 3.67PFVVEE259 pKa = 4.82EE260 pKa = 4.51GDD262 pKa = 4.1IPLQVYY268 pKa = 10.06DD269 pKa = 4.78RR270 pKa = 11.84EE271 pKa = 4.2ADD273 pKa = 3.63LEE275 pKa = 4.34FRR277 pKa = 11.84TSTPKK282 pKa = 10.52EE283 pKa = 3.67RR284 pKa = 11.84TNPIRR289 pKa = 11.84KK290 pKa = 8.87VKK292 pKa = 10.35ALYY295 pKa = 10.35NRR297 pKa = 11.84FVKK300 pKa = 9.93QVPVEE305 pKa = 4.39DD306 pKa = 4.54PLFLTAPGEE315 pKa = 3.93LVEE318 pKa = 5.86YY319 pKa = 10.45GFQNPAYY326 pKa = 9.99DD327 pKa = 3.86PDD329 pKa = 3.69EE330 pKa = 4.56TIEE333 pKa = 4.12FSIDD337 pKa = 3.11TDD339 pKa = 3.76QPTAAPDD346 pKa = 3.27SRR348 pKa = 11.84FRR350 pKa = 11.84DD351 pKa = 3.35VHH353 pKa = 4.88TLHH356 pKa = 6.74RR357 pKa = 11.84QILGEE362 pKa = 4.2APTGHH367 pKa = 5.62VRR369 pKa = 11.84VSRR372 pKa = 11.84VGSVAAMRR380 pKa = 11.84TRR382 pKa = 11.84SGATVGPQLHH392 pKa = 7.09LYY394 pKa = 10.26KK395 pKa = 10.41DD396 pKa = 3.25ISSIGEE402 pKa = 4.26AIEE405 pKa = 4.17LQPLSYY411 pKa = 10.26PHH413 pKa = 7.07ASDD416 pKa = 3.19SSTYY420 pKa = 11.43AVMPADD426 pKa = 3.56TSFGGDD432 pKa = 3.44VAVGAPPDD440 pKa = 3.43VSTEE444 pKa = 3.96VSSLEE449 pKa = 3.98EE450 pKa = 4.18GFEE453 pKa = 4.2EE454 pKa = 5.21VPLLQDD460 pKa = 4.47HH461 pKa = 7.04SGQQQTVGVSRR472 pKa = 11.84GGSRR476 pKa = 11.84DD477 pKa = 3.67RR478 pKa = 11.84QWITFEE484 pKa = 3.89IPTPRR489 pKa = 11.84TNIPASASSDD499 pKa = 2.92RR500 pKa = 11.84DD501 pKa = 3.19IFVYY505 pKa = 10.78YY506 pKa = 9.78PDD508 pKa = 4.3EE509 pKa = 4.64SQDD512 pKa = 3.37HH513 pKa = 6.89DD514 pKa = 4.38SSGEE518 pKa = 3.78AAIDD522 pKa = 3.61TDD524 pKa = 3.64ITPARR529 pKa = 11.84PIEE532 pKa = 4.64PISPVGPSYY541 pKa = 11.4VEE543 pKa = 4.15STDD546 pKa = 3.86SFPYY550 pKa = 9.38WLAPSLQRR558 pKa = 11.84KK559 pKa = 7.11KK560 pKa = 10.59RR561 pKa = 11.84KK562 pKa = 9.12RR563 pKa = 11.84KK564 pKa = 7.12TVSFCSLADD573 pKa = 3.4GCMDD577 pKa = 3.54SS578 pKa = 4.28
MM1 pKa = 7.41HH2 pKa = 6.97PAKK5 pKa = 10.15RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VPRR11 pKa = 11.84DD12 pKa = 3.01SAEE15 pKa = 3.93NIYY18 pKa = 10.51RR19 pKa = 11.84NCIHH23 pKa = 7.18FGTCPPDD30 pKa = 3.41VVNKK34 pKa = 10.4IEE36 pKa = 4.33GKK38 pKa = 8.15TVADD42 pKa = 6.06KK43 pKa = 10.81ILQWGSAGVFLGNLGISTGKK63 pKa = 10.75GSANPLGYY71 pKa = 10.54VPLGRR76 pKa = 11.84GGGVGVGAWPRR87 pKa = 11.84PGLATRR93 pKa = 11.84PVVPTAPIEE102 pKa = 4.09AVGPIDD108 pKa = 3.64VGPAVVDD115 pKa = 3.68PAVVDD120 pKa = 3.71PVLIEE125 pKa = 4.23MEE127 pKa = 5.37PILPRR132 pKa = 11.84HH133 pKa = 6.12PEE135 pKa = 3.94VVDD138 pKa = 4.48EE139 pKa = 4.74IVPIAPSDD147 pKa = 3.58PSVISDD153 pKa = 3.23NGVYY157 pKa = 9.31PAHH160 pKa = 7.05PAVIDD165 pKa = 3.84EE166 pKa = 4.43SLPTGGGRR174 pKa = 11.84TVEE177 pKa = 4.44VVAEE181 pKa = 4.07VHH183 pKa = 6.67HH184 pKa = 6.49PADD187 pKa = 4.14YY188 pKa = 10.79FPVHH192 pKa = 6.93PIDD195 pKa = 3.96TGGSDD200 pKa = 3.61NPAIIDD206 pKa = 3.63VGEE209 pKa = 5.33DD210 pKa = 3.02IPLLPRR216 pKa = 11.84TSTPHH221 pKa = 5.65HH222 pKa = 6.79TDD224 pKa = 2.94PAILHH229 pKa = 5.99ATTSFGTSAEE239 pKa = 4.27VGGARR244 pKa = 11.84YY245 pKa = 7.61TVVDD249 pKa = 5.46FEE251 pKa = 4.4ATDD254 pKa = 3.67PFVVEE259 pKa = 4.82EE260 pKa = 4.51GDD262 pKa = 4.1IPLQVYY268 pKa = 10.06DD269 pKa = 4.78RR270 pKa = 11.84EE271 pKa = 4.2ADD273 pKa = 3.63LEE275 pKa = 4.34FRR277 pKa = 11.84TSTPKK282 pKa = 10.52EE283 pKa = 3.67RR284 pKa = 11.84TNPIRR289 pKa = 11.84KK290 pKa = 8.87VKK292 pKa = 10.35ALYY295 pKa = 10.35NRR297 pKa = 11.84FVKK300 pKa = 9.93QVPVEE305 pKa = 4.39DD306 pKa = 4.54PLFLTAPGEE315 pKa = 3.93LVEE318 pKa = 5.86YY319 pKa = 10.45GFQNPAYY326 pKa = 9.99DD327 pKa = 3.86PDD329 pKa = 3.69EE330 pKa = 4.56TIEE333 pKa = 4.12FSIDD337 pKa = 3.11TDD339 pKa = 3.76QPTAAPDD346 pKa = 3.27SRR348 pKa = 11.84FRR350 pKa = 11.84DD351 pKa = 3.35VHH353 pKa = 4.88TLHH356 pKa = 6.74RR357 pKa = 11.84QILGEE362 pKa = 4.2APTGHH367 pKa = 5.62VRR369 pKa = 11.84VSRR372 pKa = 11.84VGSVAAMRR380 pKa = 11.84TRR382 pKa = 11.84SGATVGPQLHH392 pKa = 7.09LYY394 pKa = 10.26KK395 pKa = 10.41DD396 pKa = 3.25ISSIGEE402 pKa = 4.26AIEE405 pKa = 4.17LQPLSYY411 pKa = 10.26PHH413 pKa = 7.07ASDD416 pKa = 3.19SSTYY420 pKa = 11.43AVMPADD426 pKa = 3.56TSFGGDD432 pKa = 3.44VAVGAPPDD440 pKa = 3.43VSTEE444 pKa = 3.96VSSLEE449 pKa = 3.98EE450 pKa = 4.18GFEE453 pKa = 4.2EE454 pKa = 5.21VPLLQDD460 pKa = 4.47HH461 pKa = 7.04SGQQQTVGVSRR472 pKa = 11.84GGSRR476 pKa = 11.84DD477 pKa = 3.67RR478 pKa = 11.84QWITFEE484 pKa = 3.89IPTPRR489 pKa = 11.84TNIPASASSDD499 pKa = 2.92RR500 pKa = 11.84DD501 pKa = 3.19IFVYY505 pKa = 10.78YY506 pKa = 9.78PDD508 pKa = 4.3EE509 pKa = 4.64SQDD512 pKa = 3.37HH513 pKa = 6.89DD514 pKa = 4.38SSGEE518 pKa = 3.78AAIDD522 pKa = 3.61TDD524 pKa = 3.64ITPARR529 pKa = 11.84PIEE532 pKa = 4.64PISPVGPSYY541 pKa = 11.4VEE543 pKa = 4.15STDD546 pKa = 3.86SFPYY550 pKa = 9.38WLAPSLQRR558 pKa = 11.84KK559 pKa = 7.11KK560 pKa = 10.59RR561 pKa = 11.84KK562 pKa = 9.12RR563 pKa = 11.84KK564 pKa = 7.12TVSFCSLADD573 pKa = 3.4GCMDD577 pKa = 3.54SS578 pKa = 4.28
Molecular weight: 62.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P8YVV7|A0A1P8YVV7_9PAPI Regulatory protein E2 OS=Eidolon helvum papillomavirus 3 OX=1335477 GN=E2 PE=3 SV=1
MM1 pKa = 7.11EE2 pKa = 5.66ALRR5 pKa = 11.84EE6 pKa = 4.07RR7 pKa = 11.84FDD9 pKa = 3.54VLQEE13 pKa = 4.52RR14 pKa = 11.84ILKK17 pKa = 10.18HH18 pKa = 6.19YY19 pKa = 10.29EE20 pKa = 3.67LDD22 pKa = 3.77SQKK25 pKa = 10.93LADD28 pKa = 3.58HH29 pKa = 6.57CEE31 pKa = 3.49YY32 pKa = 9.96WRR34 pKa = 11.84AVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 3.68NALLHH44 pKa = 5.0YY45 pKa = 10.75ARR47 pKa = 11.84DD48 pKa = 3.8KK49 pKa = 11.01GVKK52 pKa = 8.43TLGHH56 pKa = 5.98MPVPAAQVSQARR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 4.06AIAMQLAAEE80 pKa = 4.17SLEE83 pKa = 3.96RR84 pKa = 11.84SGFGAEE90 pKa = 3.77PWSLQEE96 pKa = 4.51LSRR99 pKa = 11.84EE100 pKa = 4.41AYY102 pKa = 7.61TAAPRR107 pKa = 11.84NCFKK111 pKa = 10.97KK112 pKa = 10.08RR113 pKa = 11.84GYY115 pKa = 8.42TVLVMYY121 pKa = 10.52DD122 pKa = 3.46NSEE125 pKa = 4.32EE126 pKa = 3.98NCMEE130 pKa = 3.84YY131 pKa = 10.2TGWEE135 pKa = 4.19DD136 pKa = 4.24IYY138 pKa = 11.45VQEE141 pKa = 6.12DD142 pKa = 3.49DD143 pKa = 5.61DD144 pKa = 3.91VWLKK148 pKa = 11.26VKK150 pKa = 10.59GLVDD154 pKa = 3.76IYY156 pKa = 11.12GLYY159 pKa = 10.32YY160 pKa = 10.33EE161 pKa = 4.81SEE163 pKa = 4.28GLKK166 pKa = 10.13EE167 pKa = 4.1YY168 pKa = 10.97YY169 pKa = 8.95VTFAQDD175 pKa = 2.86AEE177 pKa = 4.84TYY179 pKa = 10.49SKK181 pKa = 9.92TGLYY185 pKa = 7.43TVKK188 pKa = 10.57YY189 pKa = 9.1KK190 pKa = 11.08SEE192 pKa = 4.05VLTNFHH198 pKa = 6.88PVSSSTPRR206 pKa = 11.84SPRR209 pKa = 11.84ARR211 pKa = 11.84RR212 pKa = 11.84QRR214 pKa = 11.84EE215 pKa = 3.97EE216 pKa = 4.33SPGSLPAGQGEE227 pKa = 4.84GEE229 pKa = 4.5GEE231 pKa = 4.47GAQTPPSKK239 pKa = 10.2RR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84GLPDD246 pKa = 2.95QGGSPTRR253 pKa = 11.84SSNPEE258 pKa = 3.75RR259 pKa = 11.84RR260 pKa = 11.84WPGQGQRR267 pKa = 11.84QGEE270 pKa = 4.65GEE272 pKa = 4.27GGRR275 pKa = 11.84PSQSPTRR282 pKa = 11.84GSAGRR287 pKa = 11.84APRR290 pKa = 11.84RR291 pKa = 11.84AARR294 pKa = 11.84SAGPGAQGRR303 pKa = 11.84GRR305 pKa = 11.84RR306 pKa = 11.84CRR308 pKa = 11.84PDD310 pKa = 3.02QAASTLAPILIVRR323 pKa = 11.84GNGNGLKK330 pKa = 9.88CLRR333 pKa = 11.84YY334 pKa = 8.9RR335 pKa = 11.84WHH337 pKa = 7.54RR338 pKa = 11.84DD339 pKa = 2.7HH340 pKa = 7.09SALFTSVSTTFRR352 pKa = 11.84WLRR355 pKa = 11.84AGLTDD360 pKa = 3.47RR361 pKa = 11.84EE362 pKa = 4.47GNAEE366 pKa = 4.07VIVRR370 pKa = 11.84FNTDD374 pKa = 2.45KK375 pKa = 10.91QRR377 pKa = 11.84EE378 pKa = 4.12DD379 pKa = 3.56VLDD382 pKa = 4.42CGAVPEE388 pKa = 4.51TVQYY392 pKa = 11.36LCGTMPFFSS401 pKa = 4.06
MM1 pKa = 7.11EE2 pKa = 5.66ALRR5 pKa = 11.84EE6 pKa = 4.07RR7 pKa = 11.84FDD9 pKa = 3.54VLQEE13 pKa = 4.52RR14 pKa = 11.84ILKK17 pKa = 10.18HH18 pKa = 6.19YY19 pKa = 10.29EE20 pKa = 3.67LDD22 pKa = 3.77SQKK25 pKa = 10.93LADD28 pKa = 3.58HH29 pKa = 6.57CEE31 pKa = 3.49YY32 pKa = 9.96WRR34 pKa = 11.84AVRR37 pKa = 11.84KK38 pKa = 9.83EE39 pKa = 3.68NALLHH44 pKa = 5.0YY45 pKa = 10.75ARR47 pKa = 11.84DD48 pKa = 3.8KK49 pKa = 11.01GVKK52 pKa = 8.43TLGHH56 pKa = 5.98MPVPAAQVSQARR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 4.06AIAMQLAAEE80 pKa = 4.17SLEE83 pKa = 3.96RR84 pKa = 11.84SGFGAEE90 pKa = 3.77PWSLQEE96 pKa = 4.51LSRR99 pKa = 11.84EE100 pKa = 4.41AYY102 pKa = 7.61TAAPRR107 pKa = 11.84NCFKK111 pKa = 10.97KK112 pKa = 10.08RR113 pKa = 11.84GYY115 pKa = 8.42TVLVMYY121 pKa = 10.52DD122 pKa = 3.46NSEE125 pKa = 4.32EE126 pKa = 3.98NCMEE130 pKa = 3.84YY131 pKa = 10.2TGWEE135 pKa = 4.19DD136 pKa = 4.24IYY138 pKa = 11.45VQEE141 pKa = 6.12DD142 pKa = 3.49DD143 pKa = 5.61DD144 pKa = 3.91VWLKK148 pKa = 11.26VKK150 pKa = 10.59GLVDD154 pKa = 3.76IYY156 pKa = 11.12GLYY159 pKa = 10.32YY160 pKa = 10.33EE161 pKa = 4.81SEE163 pKa = 4.28GLKK166 pKa = 10.13EE167 pKa = 4.1YY168 pKa = 10.97YY169 pKa = 8.95VTFAQDD175 pKa = 2.86AEE177 pKa = 4.84TYY179 pKa = 10.49SKK181 pKa = 9.92TGLYY185 pKa = 7.43TVKK188 pKa = 10.57YY189 pKa = 9.1KK190 pKa = 11.08SEE192 pKa = 4.05VLTNFHH198 pKa = 6.88PVSSSTPRR206 pKa = 11.84SPRR209 pKa = 11.84ARR211 pKa = 11.84RR212 pKa = 11.84QRR214 pKa = 11.84EE215 pKa = 3.97EE216 pKa = 4.33SPGSLPAGQGEE227 pKa = 4.84GEE229 pKa = 4.5GEE231 pKa = 4.47GAQTPPSKK239 pKa = 10.2RR240 pKa = 11.84RR241 pKa = 11.84RR242 pKa = 11.84GLPDD246 pKa = 2.95QGGSPTRR253 pKa = 11.84SSNPEE258 pKa = 3.75RR259 pKa = 11.84RR260 pKa = 11.84WPGQGQRR267 pKa = 11.84QGEE270 pKa = 4.65GEE272 pKa = 4.27GGRR275 pKa = 11.84PSQSPTRR282 pKa = 11.84GSAGRR287 pKa = 11.84APRR290 pKa = 11.84RR291 pKa = 11.84AARR294 pKa = 11.84SAGPGAQGRR303 pKa = 11.84GRR305 pKa = 11.84RR306 pKa = 11.84CRR308 pKa = 11.84PDD310 pKa = 3.02QAASTLAPILIVRR323 pKa = 11.84GNGNGLKK330 pKa = 9.88CLRR333 pKa = 11.84YY334 pKa = 8.9RR335 pKa = 11.84WHH337 pKa = 7.54RR338 pKa = 11.84DD339 pKa = 2.7HH340 pKa = 7.09SALFTSVSTTFRR352 pKa = 11.84WLRR355 pKa = 11.84AGLTDD360 pKa = 3.47RR361 pKa = 11.84EE362 pKa = 4.47GNAEE366 pKa = 4.07VIVRR370 pKa = 11.84FNTDD374 pKa = 2.45KK375 pKa = 10.91QRR377 pKa = 11.84EE378 pKa = 4.12DD379 pKa = 3.56VLDD382 pKa = 4.42CGAVPEE388 pKa = 4.51TVQYY392 pKa = 11.36LCGTMPFFSS401 pKa = 4.06
Molecular weight: 45.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2415 |
87 |
660 |
402.5 |
44.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.578 ± 0.431 | 2.195 ± 0.62 |
6.418 ± 0.472 | 6.625 ± 0.619 |
3.106 ± 0.278 | 7.246 ± 0.561 |
2.153 ± 0.241 | 4.265 ± 0.634 |
4.224 ± 0.596 | 7.495 ± 0.855 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.029 ± 0.416 | 3.271 ± 0.463 |
6.791 ± 1.204 | 3.644 ± 0.449 |
7.081 ± 0.876 | 7.039 ± 0.299 |
6.791 ± 0.641 | 7.288 ± 0.887 |
1.491 ± 0.24 | 3.271 ± 0.334 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
