
Desulfovibrio sp. 6_1_46AFAA
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; unclassified Desulfovibrio
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
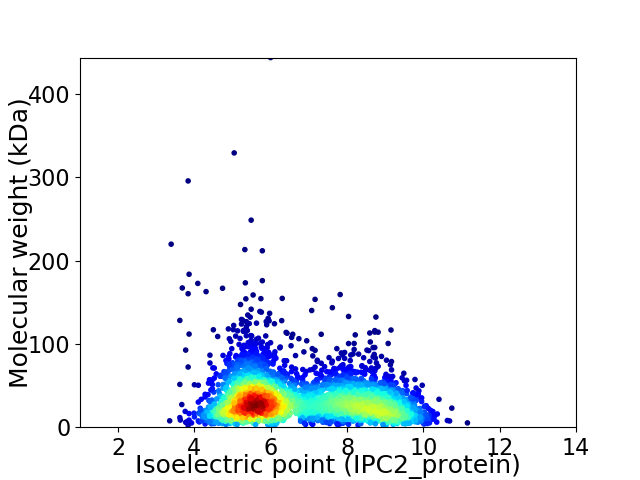
Virtual 2D-PAGE plot for 3312 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G1UPV3|G1UPV3_9DELT Uncharacterized protein OS=Desulfovibrio sp. 6_1_46AFAA OX=665942 GN=HMPREF1022_00626 PE=4 SV=1
MM1 pKa = 7.28PTTHH5 pKa = 7.29ILHH8 pKa = 6.75LPAAGQKK15 pKa = 8.43TVLAGFSPQSRR26 pKa = 11.84MVLDD30 pKa = 4.44FPTRR34 pKa = 11.84GVFAGRR40 pKa = 11.84EE41 pKa = 4.22GNDD44 pKa = 3.33LVFSRR49 pKa = 11.84IDD51 pKa = 3.26GTRR54 pKa = 11.84LVLKK58 pKa = 10.12DD59 pKa = 3.05YY60 pKa = 9.81HH61 pKa = 7.16APQNVPEE68 pKa = 5.09DD69 pKa = 3.98VPQNAPADD77 pKa = 3.79GAPRR81 pKa = 11.84DD82 pKa = 4.04EE83 pKa = 4.88GPPAIVVDD91 pKa = 4.26GRR93 pKa = 11.84EE94 pKa = 3.99MTHH97 pKa = 6.01QAFFDD102 pKa = 3.77ALGEE106 pKa = 4.22DD107 pKa = 4.82DD108 pKa = 4.59MPQAGVVQARR118 pKa = 11.84NSAYY122 pKa = 10.01HH123 pKa = 6.73EE124 pKa = 4.25YY125 pKa = 10.72SEE127 pKa = 5.16SEE129 pKa = 3.93LSKK132 pKa = 11.18GISEE136 pKa = 5.03LSALDD141 pKa = 3.6DD142 pKa = 4.74HH143 pKa = 6.14EE144 pKa = 5.0TEE146 pKa = 4.46SDD148 pKa = 3.52TNKK151 pKa = 10.2PLGSRR156 pKa = 11.84EE157 pKa = 3.61GRR159 pKa = 11.84QDD161 pKa = 3.2DD162 pKa = 4.1FAAEE166 pKa = 4.03RR167 pKa = 11.84GGVLGGTDD175 pKa = 3.4TAGSGSNGSGSGSPGGSQGGNGGSGGSGHH204 pKa = 6.61GGSGGTGGTGGNGSGGTGGTGNSNPSVASATVTIAEE240 pKa = 4.31DD241 pKa = 3.98TISVSGALPSPTDD254 pKa = 3.77PNAGDD259 pKa = 3.42ILSFIAKK266 pKa = 9.86ANAAGLYY273 pKa = 7.61GTLSVDD279 pKa = 3.08ANGNYY284 pKa = 9.6IYY286 pKa = 10.41ILNNALSAVQGLGVGEE302 pKa = 4.65SLTDD306 pKa = 3.28TFTFTVTDD314 pKa = 3.67NHH316 pKa = 6.75GGTANNTLTVTITGTNDD333 pKa = 3.1APVLAATTVNVSEE346 pKa = 5.08DD347 pKa = 3.61SNITSGILPTPTDD360 pKa = 3.24VDD362 pKa = 3.68VHH364 pKa = 6.46DD365 pKa = 4.29VVSFVPQTNTAGLYY379 pKa = 7.68GTLSVDD385 pKa = 3.05ANGNYY390 pKa = 9.81TYY392 pKa = 11.26VLNNTLPAVQGLGAGEE408 pKa = 4.41NLTDD412 pKa = 3.65TFTVTVTDD420 pKa = 3.52NHH422 pKa = 6.7GGTSSAVVTVNINGSNDD439 pKa = 3.23APIVSAVATSIFEE452 pKa = 4.27DD453 pKa = 3.79TASVSGTLPAPVDD466 pKa = 3.8PDD468 pKa = 3.47LHH470 pKa = 7.15DD471 pKa = 3.73VASYY475 pKa = 10.86LPQTNTVGLYY485 pKa = 7.99GTLTLDD491 pKa = 3.08ASGNYY496 pKa = 9.26TYY498 pKa = 10.83TLNNSLSAVQGLGVGEE514 pKa = 4.65SLTDD518 pKa = 3.3TFTFTASDD526 pKa = 3.55GHH528 pKa = 6.58GGTSSSTLTVTINGSNDD545 pKa = 3.3APVLAATVVGVDD557 pKa = 3.58KK558 pKa = 11.29DD559 pKa = 4.21DD560 pKa = 4.64VITNGTLPTPTDD572 pKa = 3.22VDD574 pKa = 3.63VHH576 pKa = 6.17DD577 pKa = 4.46TVSFTPLTNAPGSYY591 pKa = 9.34GTFSVDD597 pKa = 2.89ANGNYY602 pKa = 9.77TYY604 pKa = 11.3VLNTSLPAVQGLGPADD620 pKa = 3.91SLTDD624 pKa = 3.51TFTVTVTDD632 pKa = 3.32NHH634 pKa = 6.35GASSSTVITVSINGSSNDD652 pKa = 3.5APTVAAALASIGEE665 pKa = 4.4DD666 pKa = 3.54TPSVTGVLPTPGDD679 pKa = 4.15PDD681 pKa = 3.45PHH683 pKa = 7.65DD684 pKa = 3.45VVTFVPQAGTAGLYY698 pKa = 8.57GTFSVDD704 pKa = 2.64ASGNYY709 pKa = 9.32TYY711 pKa = 10.74TLNNSLPAVQGLGAGEE727 pKa = 4.47SLTEE731 pKa = 4.05TFTFTVSDD739 pKa = 3.8GRR741 pKa = 11.84GGTASNTLTVTIDD754 pKa = 3.61GSNDD758 pKa = 3.22APVVTAATASIVEE771 pKa = 4.26DD772 pKa = 3.37TAGISGTLPPAVDD785 pKa = 3.93PDD787 pKa = 3.71LHH789 pKa = 7.18DD790 pKa = 3.6VATFIPQTNTAGLYY804 pKa = 7.74GTLTLDD810 pKa = 3.08ASGNYY815 pKa = 9.26TYY817 pKa = 10.74TLNNSLPAVQGLGVGEE833 pKa = 4.56SLTEE837 pKa = 4.12TFTFTATDD845 pKa = 3.44GHH847 pKa = 6.75GGTSSNTLTVTINGSNDD864 pKa = 3.3APVLAATTVNVAEE877 pKa = 5.0DD878 pKa = 3.71SAVTTGTLPMPTDD891 pKa = 3.4VDD893 pKa = 3.73VHH895 pKa = 6.18DD896 pKa = 4.44TVSFAPLTNAPGNYY910 pKa = 8.29GTFSVDD916 pKa = 2.85ANGNYY921 pKa = 9.77TYY923 pKa = 11.3VLNTSLPTVQGLGAGEE939 pKa = 4.61SLTDD943 pKa = 3.48SFTVTVSDD951 pKa = 3.4NHH953 pKa = 6.57GGSSSTVITVSINGSNDD970 pKa = 2.9APTVSAAATAIDD982 pKa = 4.59EE983 pKa = 4.53DD984 pKa = 4.37TASVSGTLPVPVDD997 pKa = 3.71PDD999 pKa = 3.31LHH1001 pKa = 7.22DD1002 pKa = 3.67VVSFVPQTNAPGGYY1016 pKa = 7.97GTLSVDD1022 pKa = 2.91ASGNYY1027 pKa = 9.32TYY1029 pKa = 10.74TLNNSLPAVQALGVGEE1045 pKa = 4.66SLTDD1049 pKa = 3.28TFTFTVTDD1057 pKa = 3.35NHH1059 pKa = 6.79GATTSNTLTVTINGTNDD1076 pKa = 3.28LPTVSAAAASIAEE1089 pKa = 4.23DD1090 pKa = 3.5TAGISGMLPTPVDD1103 pKa = 3.4VDD1105 pKa = 3.56VHH1107 pKa = 5.85DD1108 pKa = 4.34TVTFTPQTNTAGLYY1122 pKa = 7.69GTLNVDD1128 pKa = 2.84ADD1130 pKa = 3.86GNYY1133 pKa = 10.25TYY1135 pKa = 10.45TLNNASPAVQGLGVGEE1151 pKa = 4.51NLTDD1155 pKa = 3.71TFPFTVSDD1163 pKa = 3.69GHH1165 pKa = 6.83GGTATNTLTVTINGTNDD1182 pKa = 3.11APIVAAATTAIAEE1195 pKa = 4.32DD1196 pKa = 3.74TAGVSGILPMPTDD1209 pKa = 3.43VDD1211 pKa = 3.73VHH1213 pKa = 6.17DD1214 pKa = 4.49TVSFTPQTNAPGSYY1228 pKa = 9.24GTFSVDD1234 pKa = 3.01ADD1236 pKa = 3.35GHH1238 pKa = 4.49YY1239 pKa = 9.84TYY1241 pKa = 10.52TLNNALPAVQGLGAGEE1257 pKa = 4.59SLTDD1261 pKa = 3.26TFTFTVSDD1269 pKa = 3.65GRR1271 pKa = 11.84GARR1274 pKa = 11.84APIPP1278 pKa = 3.75
MM1 pKa = 7.28PTTHH5 pKa = 7.29ILHH8 pKa = 6.75LPAAGQKK15 pKa = 8.43TVLAGFSPQSRR26 pKa = 11.84MVLDD30 pKa = 4.44FPTRR34 pKa = 11.84GVFAGRR40 pKa = 11.84EE41 pKa = 4.22GNDD44 pKa = 3.33LVFSRR49 pKa = 11.84IDD51 pKa = 3.26GTRR54 pKa = 11.84LVLKK58 pKa = 10.12DD59 pKa = 3.05YY60 pKa = 9.81HH61 pKa = 7.16APQNVPEE68 pKa = 5.09DD69 pKa = 3.98VPQNAPADD77 pKa = 3.79GAPRR81 pKa = 11.84DD82 pKa = 4.04EE83 pKa = 4.88GPPAIVVDD91 pKa = 4.26GRR93 pKa = 11.84EE94 pKa = 3.99MTHH97 pKa = 6.01QAFFDD102 pKa = 3.77ALGEE106 pKa = 4.22DD107 pKa = 4.82DD108 pKa = 4.59MPQAGVVQARR118 pKa = 11.84NSAYY122 pKa = 10.01HH123 pKa = 6.73EE124 pKa = 4.25YY125 pKa = 10.72SEE127 pKa = 5.16SEE129 pKa = 3.93LSKK132 pKa = 11.18GISEE136 pKa = 5.03LSALDD141 pKa = 3.6DD142 pKa = 4.74HH143 pKa = 6.14EE144 pKa = 5.0TEE146 pKa = 4.46SDD148 pKa = 3.52TNKK151 pKa = 10.2PLGSRR156 pKa = 11.84EE157 pKa = 3.61GRR159 pKa = 11.84QDD161 pKa = 3.2DD162 pKa = 4.1FAAEE166 pKa = 4.03RR167 pKa = 11.84GGVLGGTDD175 pKa = 3.4TAGSGSNGSGSGSPGGSQGGNGGSGGSGHH204 pKa = 6.61GGSGGTGGTGGNGSGGTGGTGNSNPSVASATVTIAEE240 pKa = 4.31DD241 pKa = 3.98TISVSGALPSPTDD254 pKa = 3.77PNAGDD259 pKa = 3.42ILSFIAKK266 pKa = 9.86ANAAGLYY273 pKa = 7.61GTLSVDD279 pKa = 3.08ANGNYY284 pKa = 9.6IYY286 pKa = 10.41ILNNALSAVQGLGVGEE302 pKa = 4.65SLTDD306 pKa = 3.28TFTFTVTDD314 pKa = 3.67NHH316 pKa = 6.75GGTANNTLTVTITGTNDD333 pKa = 3.1APVLAATTVNVSEE346 pKa = 5.08DD347 pKa = 3.61SNITSGILPTPTDD360 pKa = 3.24VDD362 pKa = 3.68VHH364 pKa = 6.46DD365 pKa = 4.29VVSFVPQTNTAGLYY379 pKa = 7.68GTLSVDD385 pKa = 3.05ANGNYY390 pKa = 9.81TYY392 pKa = 11.26VLNNTLPAVQGLGAGEE408 pKa = 4.41NLTDD412 pKa = 3.65TFTVTVTDD420 pKa = 3.52NHH422 pKa = 6.7GGTSSAVVTVNINGSNDD439 pKa = 3.23APIVSAVATSIFEE452 pKa = 4.27DD453 pKa = 3.79TASVSGTLPAPVDD466 pKa = 3.8PDD468 pKa = 3.47LHH470 pKa = 7.15DD471 pKa = 3.73VASYY475 pKa = 10.86LPQTNTVGLYY485 pKa = 7.99GTLTLDD491 pKa = 3.08ASGNYY496 pKa = 9.26TYY498 pKa = 10.83TLNNSLSAVQGLGVGEE514 pKa = 4.65SLTDD518 pKa = 3.3TFTFTASDD526 pKa = 3.55GHH528 pKa = 6.58GGTSSSTLTVTINGSNDD545 pKa = 3.3APVLAATVVGVDD557 pKa = 3.58KK558 pKa = 11.29DD559 pKa = 4.21DD560 pKa = 4.64VITNGTLPTPTDD572 pKa = 3.22VDD574 pKa = 3.63VHH576 pKa = 6.17DD577 pKa = 4.46TVSFTPLTNAPGSYY591 pKa = 9.34GTFSVDD597 pKa = 2.89ANGNYY602 pKa = 9.77TYY604 pKa = 11.3VLNTSLPAVQGLGPADD620 pKa = 3.91SLTDD624 pKa = 3.51TFTVTVTDD632 pKa = 3.32NHH634 pKa = 6.35GASSSTVITVSINGSSNDD652 pKa = 3.5APTVAAALASIGEE665 pKa = 4.4DD666 pKa = 3.54TPSVTGVLPTPGDD679 pKa = 4.15PDD681 pKa = 3.45PHH683 pKa = 7.65DD684 pKa = 3.45VVTFVPQAGTAGLYY698 pKa = 8.57GTFSVDD704 pKa = 2.64ASGNYY709 pKa = 9.32TYY711 pKa = 10.74TLNNSLPAVQGLGAGEE727 pKa = 4.47SLTEE731 pKa = 4.05TFTFTVSDD739 pKa = 3.8GRR741 pKa = 11.84GGTASNTLTVTIDD754 pKa = 3.61GSNDD758 pKa = 3.22APVVTAATASIVEE771 pKa = 4.26DD772 pKa = 3.37TAGISGTLPPAVDD785 pKa = 3.93PDD787 pKa = 3.71LHH789 pKa = 7.18DD790 pKa = 3.6VATFIPQTNTAGLYY804 pKa = 7.74GTLTLDD810 pKa = 3.08ASGNYY815 pKa = 9.26TYY817 pKa = 10.74TLNNSLPAVQGLGVGEE833 pKa = 4.56SLTEE837 pKa = 4.12TFTFTATDD845 pKa = 3.44GHH847 pKa = 6.75GGTSSNTLTVTINGSNDD864 pKa = 3.3APVLAATTVNVAEE877 pKa = 5.0DD878 pKa = 3.71SAVTTGTLPMPTDD891 pKa = 3.4VDD893 pKa = 3.73VHH895 pKa = 6.18DD896 pKa = 4.44TVSFAPLTNAPGNYY910 pKa = 8.29GTFSVDD916 pKa = 2.85ANGNYY921 pKa = 9.77TYY923 pKa = 11.3VLNTSLPTVQGLGAGEE939 pKa = 4.61SLTDD943 pKa = 3.48SFTVTVSDD951 pKa = 3.4NHH953 pKa = 6.57GGSSSTVITVSINGSNDD970 pKa = 2.9APTVSAAATAIDD982 pKa = 4.59EE983 pKa = 4.53DD984 pKa = 4.37TASVSGTLPVPVDD997 pKa = 3.71PDD999 pKa = 3.31LHH1001 pKa = 7.22DD1002 pKa = 3.67VVSFVPQTNAPGGYY1016 pKa = 7.97GTLSVDD1022 pKa = 2.91ASGNYY1027 pKa = 9.32TYY1029 pKa = 10.74TLNNSLPAVQALGVGEE1045 pKa = 4.66SLTDD1049 pKa = 3.28TFTFTVTDD1057 pKa = 3.35NHH1059 pKa = 6.79GATTSNTLTVTINGTNDD1076 pKa = 3.28LPTVSAAAASIAEE1089 pKa = 4.23DD1090 pKa = 3.5TAGISGMLPTPVDD1103 pKa = 3.4VDD1105 pKa = 3.56VHH1107 pKa = 5.85DD1108 pKa = 4.34TVTFTPQTNTAGLYY1122 pKa = 7.69GTLNVDD1128 pKa = 2.84ADD1130 pKa = 3.86GNYY1133 pKa = 10.25TYY1135 pKa = 10.45TLNNASPAVQGLGVGEE1151 pKa = 4.51NLTDD1155 pKa = 3.71TFPFTVSDD1163 pKa = 3.69GHH1165 pKa = 6.83GGTATNTLTVTINGTNDD1182 pKa = 3.11APIVAAATTAIAEE1195 pKa = 4.32DD1196 pKa = 3.74TAGVSGILPMPTDD1209 pKa = 3.43VDD1211 pKa = 3.73VHH1213 pKa = 6.17DD1214 pKa = 4.49TVSFTPQTNAPGSYY1228 pKa = 9.24GTFSVDD1234 pKa = 3.01ADD1236 pKa = 3.35GHH1238 pKa = 4.49YY1239 pKa = 9.84TYY1241 pKa = 10.52TLNNALPAVQGLGAGEE1257 pKa = 4.59SLTDD1261 pKa = 3.26TFTFTVSDD1269 pKa = 3.65GRR1271 pKa = 11.84GARR1274 pKa = 11.84APIPP1278 pKa = 3.75
Molecular weight: 128.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G1UTI6|G1UTI6_9DELT GDP-mannose 4 6-dehydratase OS=Desulfovibrio sp. 6_1_46AFAA OX=665942 GN=gmd PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.07IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPSGRR28 pKa = 11.84AILRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.57NLSAA44 pKa = 4.67
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.07QPSKK9 pKa = 9.07IRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.96GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPSGRR28 pKa = 11.84AILRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.57NLSAA44 pKa = 4.67
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1086973 |
22 |
4049 |
328.2 |
35.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.522 ± 0.056 | 1.518 ± 0.023 |
5.245 ± 0.037 | 6.093 ± 0.049 |
3.699 ± 0.027 | 8.276 ± 0.044 |
2.03 ± 0.022 | 4.477 ± 0.04 |
3.839 ± 0.042 | 11.166 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.594 ± 0.02 | 3.005 ± 0.034 |
5.223 ± 0.04 | 3.363 ± 0.026 |
6.984 ± 0.056 | 5.456 ± 0.03 |
4.802 ± 0.052 | 6.859 ± 0.034 |
1.318 ± 0.018 | 2.531 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
