
Tolypocladium cylindrosporum virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
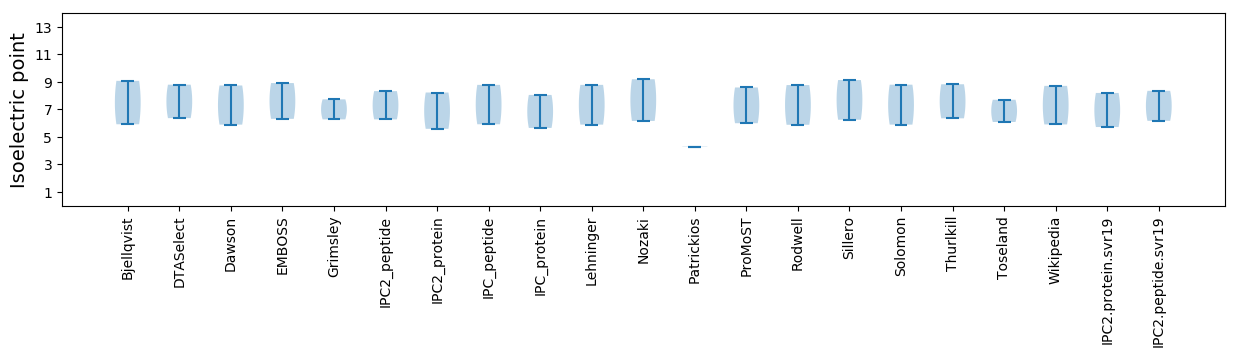
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
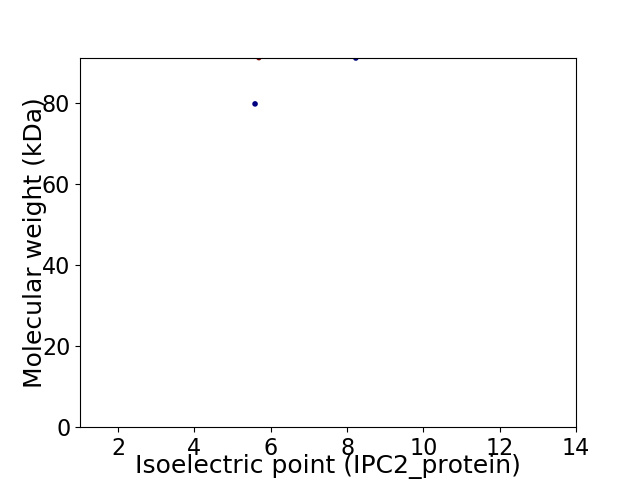
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7BBP7|E7BBP7_9VIRU RNA-directed RNA polymerase OS=Tolypocladium cylindrosporum virus 1 OX=939923 GN=RdRp PE=3 SV=1
MM1 pKa = 7.86DD2 pKa = 5.02FIIRR6 pKa = 11.84NSFLSGVIASPRR18 pKa = 11.84GGSLDD23 pKa = 3.1NDD25 pKa = 3.12NRR27 pKa = 11.84FRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 7.37RR32 pKa = 11.84TSVRR36 pKa = 11.84TTATIGGNDD45 pKa = 3.86DD46 pKa = 3.68SRR48 pKa = 11.84TTSIFYY54 pKa = 10.38EE55 pKa = 4.05VGRR58 pKa = 11.84AVNTKK63 pKa = 9.25GRR65 pKa = 11.84ALNRR69 pKa = 11.84PSDD72 pKa = 3.56GALCIEE78 pKa = 4.64AAYY81 pKa = 7.13PTNTVLAEE89 pKa = 4.09DD90 pKa = 5.26FIGLAKK96 pKa = 10.54KK97 pKa = 8.47YY98 pKa = 8.69TNFSASFEE106 pKa = 4.23YY107 pKa = 10.81SSLAGVAEE115 pKa = 4.06RR116 pKa = 11.84LARR119 pKa = 11.84ALAASSVFTDD129 pKa = 3.16VDD131 pKa = 4.2SNDD134 pKa = 2.92IRR136 pKa = 11.84GGAGLVVNAVGTYY149 pKa = 9.9DD150 pKa = 4.05GPISSLTNTVYY161 pKa = 10.38IPRR164 pKa = 11.84LVNSSITGEE173 pKa = 4.26VFSVLANAVSGEE185 pKa = 4.15GSSVATDD192 pKa = 4.25IIEE195 pKa = 4.41LDD197 pKa = 3.31AGTRR201 pKa = 11.84QPLIPEE207 pKa = 4.07VDD209 pKa = 3.63PAGLAAACVDD219 pKa = 3.5ALRR222 pKa = 11.84IVGSNMIASDD232 pKa = 3.62QGPLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 5.82RR247 pKa = 11.84VLSVVGHH254 pKa = 5.45TDD256 pKa = 2.62EE257 pKa = 5.93GGITRR262 pKa = 11.84DD263 pKa = 3.64LLRR266 pKa = 11.84CSSFAPPFGGIHH278 pKa = 6.0YY279 pKa = 10.37ALEE282 pKa = 4.83PYY284 pKa = 10.56AGLPALQFTAFPAYY298 pKa = 9.82AAYY301 pKa = 10.32VDD303 pKa = 5.23SIALTTAAVVAHH315 pKa = 7.19ADD317 pKa = 3.45PGVMYY322 pKa = 10.42GGSWFPTFYY331 pKa = 11.02NGTADD336 pKa = 2.9SDD338 pKa = 4.01GTVRR342 pKa = 11.84PGQNQAGTAAMANRR356 pKa = 11.84NRR358 pKa = 11.84AQLLGSAPAFFSEE371 pKa = 4.6YY372 pKa = 10.01IRR374 pKa = 11.84GLATIFGASGDD385 pKa = 3.63SSLACRR391 pKa = 11.84FMGAASYY398 pKa = 10.82ALPQDD403 pKa = 3.91PRR405 pKa = 11.84HH406 pKa = 5.75LRR408 pKa = 11.84YY409 pKa = 8.93ATVSPWFWIEE419 pKa = 3.69PTGLLPHH426 pKa = 7.31DD427 pKa = 4.03FLGSKK432 pKa = 10.63AEE434 pKa = 4.11ANGAGSFAWKK444 pKa = 8.74DD445 pKa = 3.4TTRR448 pKa = 11.84TRR450 pKa = 11.84VAWDD454 pKa = 3.53EE455 pKa = 4.01LTQHH459 pKa = 6.61GEE461 pKa = 3.64ADD463 pKa = 3.95TTFSAYY469 pKa = 8.54VARR472 pKa = 11.84FRR474 pKa = 11.84SPRR477 pKa = 11.84QQWFFAHH484 pKa = 6.75WMNHH488 pKa = 4.98PLNGLGAIRR497 pKa = 11.84VRR499 pKa = 11.84QLDD502 pKa = 3.58PNGIVQPGQCVAHH515 pKa = 7.03PDD517 pKa = 3.13VRR519 pKa = 11.84DD520 pKa = 3.42RR521 pKa = 11.84VEE523 pKa = 4.61ADD525 pKa = 4.14LPFTDD530 pKa = 4.54YY531 pKa = 11.29LWTRR535 pKa = 11.84GQSPPPAPGEE545 pKa = 3.87LLNLSGSVGFLVRR558 pKa = 11.84HH559 pKa = 5.7LTYY562 pKa = 11.09DD563 pKa = 3.76DD564 pKa = 5.25DD565 pKa = 4.58GVPHH569 pKa = 6.99EE570 pKa = 4.37EE571 pKa = 4.0HH572 pKa = 6.37VPTRR576 pKa = 11.84RR577 pKa = 11.84EE578 pKa = 3.94FLDD581 pKa = 3.35TTVTIEE587 pKa = 3.5VGRR590 pKa = 11.84PVGIATGASNAGDD603 pKa = 3.29NHH605 pKa = 5.37VRR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TKK612 pKa = 10.34AANEE616 pKa = 3.42LAAARR621 pKa = 11.84RR622 pKa = 11.84RR623 pKa = 11.84AAVFGRR629 pKa = 11.84ADD631 pKa = 3.47VAEE634 pKa = 4.15MPILTSAPALAPPPSAGLEE653 pKa = 3.97RR654 pKa = 11.84GSDD657 pKa = 3.54PGGSGSQRR665 pKa = 11.84RR666 pKa = 11.84AVPAGLPGNDD676 pKa = 2.51TWARR680 pKa = 11.84EE681 pKa = 4.11PTGAPRR687 pKa = 11.84NPVPHH692 pKa = 6.4HH693 pKa = 5.9QPLRR697 pKa = 11.84APQLARR703 pKa = 11.84QAGGLGGGAAPIPPPPPGVPAPPALPPADD732 pKa = 5.29DD733 pKa = 5.48DD734 pKa = 4.3NAPPAPVATAPPVADD749 pKa = 3.92PQAMVGDD756 pKa = 4.23QII758 pKa = 4.63
MM1 pKa = 7.86DD2 pKa = 5.02FIIRR6 pKa = 11.84NSFLSGVIASPRR18 pKa = 11.84GGSLDD23 pKa = 3.1NDD25 pKa = 3.12NRR27 pKa = 11.84FRR29 pKa = 11.84RR30 pKa = 11.84YY31 pKa = 7.37RR32 pKa = 11.84TSVRR36 pKa = 11.84TTATIGGNDD45 pKa = 3.86DD46 pKa = 3.68SRR48 pKa = 11.84TTSIFYY54 pKa = 10.38EE55 pKa = 4.05VGRR58 pKa = 11.84AVNTKK63 pKa = 9.25GRR65 pKa = 11.84ALNRR69 pKa = 11.84PSDD72 pKa = 3.56GALCIEE78 pKa = 4.64AAYY81 pKa = 7.13PTNTVLAEE89 pKa = 4.09DD90 pKa = 5.26FIGLAKK96 pKa = 10.54KK97 pKa = 8.47YY98 pKa = 8.69TNFSASFEE106 pKa = 4.23YY107 pKa = 10.81SSLAGVAEE115 pKa = 4.06RR116 pKa = 11.84LARR119 pKa = 11.84ALAASSVFTDD129 pKa = 3.16VDD131 pKa = 4.2SNDD134 pKa = 2.92IRR136 pKa = 11.84GGAGLVVNAVGTYY149 pKa = 9.9DD150 pKa = 4.05GPISSLTNTVYY161 pKa = 10.38IPRR164 pKa = 11.84LVNSSITGEE173 pKa = 4.26VFSVLANAVSGEE185 pKa = 4.15GSSVATDD192 pKa = 4.25IIEE195 pKa = 4.41LDD197 pKa = 3.31AGTRR201 pKa = 11.84QPLIPEE207 pKa = 4.07VDD209 pKa = 3.63PAGLAAACVDD219 pKa = 3.5ALRR222 pKa = 11.84IVGSNMIASDD232 pKa = 3.62QGPLFSLAVTRR243 pKa = 11.84GIHH246 pKa = 5.82RR247 pKa = 11.84VLSVVGHH254 pKa = 5.45TDD256 pKa = 2.62EE257 pKa = 5.93GGITRR262 pKa = 11.84DD263 pKa = 3.64LLRR266 pKa = 11.84CSSFAPPFGGIHH278 pKa = 6.0YY279 pKa = 10.37ALEE282 pKa = 4.83PYY284 pKa = 10.56AGLPALQFTAFPAYY298 pKa = 9.82AAYY301 pKa = 10.32VDD303 pKa = 5.23SIALTTAAVVAHH315 pKa = 7.19ADD317 pKa = 3.45PGVMYY322 pKa = 10.42GGSWFPTFYY331 pKa = 11.02NGTADD336 pKa = 2.9SDD338 pKa = 4.01GTVRR342 pKa = 11.84PGQNQAGTAAMANRR356 pKa = 11.84NRR358 pKa = 11.84AQLLGSAPAFFSEE371 pKa = 4.6YY372 pKa = 10.01IRR374 pKa = 11.84GLATIFGASGDD385 pKa = 3.63SSLACRR391 pKa = 11.84FMGAASYY398 pKa = 10.82ALPQDD403 pKa = 3.91PRR405 pKa = 11.84HH406 pKa = 5.75LRR408 pKa = 11.84YY409 pKa = 8.93ATVSPWFWIEE419 pKa = 3.69PTGLLPHH426 pKa = 7.31DD427 pKa = 4.03FLGSKK432 pKa = 10.63AEE434 pKa = 4.11ANGAGSFAWKK444 pKa = 8.74DD445 pKa = 3.4TTRR448 pKa = 11.84TRR450 pKa = 11.84VAWDD454 pKa = 3.53EE455 pKa = 4.01LTQHH459 pKa = 6.61GEE461 pKa = 3.64ADD463 pKa = 3.95TTFSAYY469 pKa = 8.54VARR472 pKa = 11.84FRR474 pKa = 11.84SPRR477 pKa = 11.84QQWFFAHH484 pKa = 6.75WMNHH488 pKa = 4.98PLNGLGAIRR497 pKa = 11.84VRR499 pKa = 11.84QLDD502 pKa = 3.58PNGIVQPGQCVAHH515 pKa = 7.03PDD517 pKa = 3.13VRR519 pKa = 11.84DD520 pKa = 3.42RR521 pKa = 11.84VEE523 pKa = 4.61ADD525 pKa = 4.14LPFTDD530 pKa = 4.54YY531 pKa = 11.29LWTRR535 pKa = 11.84GQSPPPAPGEE545 pKa = 3.87LLNLSGSVGFLVRR558 pKa = 11.84HH559 pKa = 5.7LTYY562 pKa = 11.09DD563 pKa = 3.76DD564 pKa = 5.25DD565 pKa = 4.58GVPHH569 pKa = 6.99EE570 pKa = 4.37EE571 pKa = 4.0HH572 pKa = 6.37VPTRR576 pKa = 11.84RR577 pKa = 11.84EE578 pKa = 3.94FLDD581 pKa = 3.35TTVTIEE587 pKa = 3.5VGRR590 pKa = 11.84PVGIATGASNAGDD603 pKa = 3.29NHH605 pKa = 5.37VRR607 pKa = 11.84RR608 pKa = 11.84ARR610 pKa = 11.84TKK612 pKa = 10.34AANEE616 pKa = 3.42LAAARR621 pKa = 11.84RR622 pKa = 11.84RR623 pKa = 11.84AAVFGRR629 pKa = 11.84ADD631 pKa = 3.47VAEE634 pKa = 4.15MPILTSAPALAPPPSAGLEE653 pKa = 3.97RR654 pKa = 11.84GSDD657 pKa = 3.54PGGSGSQRR665 pKa = 11.84RR666 pKa = 11.84AVPAGLPGNDD676 pKa = 2.51TWARR680 pKa = 11.84EE681 pKa = 4.11PTGAPRR687 pKa = 11.84NPVPHH692 pKa = 6.4HH693 pKa = 5.9QPLRR697 pKa = 11.84APQLARR703 pKa = 11.84QAGGLGGGAAPIPPPPPGVPAPPALPPADD732 pKa = 5.29DD733 pKa = 5.48DD734 pKa = 4.3NAPPAPVATAPPVADD749 pKa = 3.92PQAMVGDD756 pKa = 4.23QII758 pKa = 4.63
Molecular weight: 79.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7BBP7|E7BBP7_9VIRU RNA-directed RNA polymerase OS=Tolypocladium cylindrosporum virus 1 OX=939923 GN=RdRp PE=3 SV=1
MM1 pKa = 7.13VFVSGRR7 pKa = 11.84VGEE10 pKa = 4.65FGALGSYY17 pKa = 10.26LHH19 pKa = 7.35DD20 pKa = 5.03LLPEE24 pKa = 4.86LGDD27 pKa = 3.62VTEE30 pKa = 5.08SYY32 pKa = 11.86ADD34 pKa = 3.84LEE36 pKa = 4.35LTEE39 pKa = 4.14QLIKK43 pKa = 10.5FATHH47 pKa = 5.6SVSLRR52 pKa = 11.84GVHH55 pKa = 6.25PLGPVAVSLLCVAYY69 pKa = 9.08PVQCDD74 pKa = 3.76FRR76 pKa = 11.84LSDD79 pKa = 3.54LTAVLRR85 pKa = 11.84HH86 pKa = 5.78SFSFLPQRR94 pKa = 11.84APLLEE99 pKa = 3.99LTGLIEE105 pKa = 4.77RR106 pKa = 11.84RR107 pKa = 11.84VTLRR111 pKa = 11.84KK112 pKa = 9.91APATIGALHH121 pKa = 6.17TKK123 pKa = 8.48VARR126 pKa = 11.84LCNSDD131 pKa = 3.11SAARR135 pKa = 11.84SQLWPPKK142 pKa = 8.94RR143 pKa = 11.84HH144 pKa = 5.7AAAGTKK150 pKa = 10.25VNIRR154 pKa = 11.84LAPLAVALSRR164 pKa = 11.84SYY166 pKa = 11.34GPAWLGRR173 pKa = 11.84ALTPLVGCAEE183 pKa = 4.38DD184 pKa = 4.45AVCCALLLATALEE197 pKa = 4.21PHH199 pKa = 6.84FGHH202 pKa = 7.42LGLEE206 pKa = 4.79VAQSMILEE214 pKa = 4.35PNNAKK219 pKa = 10.3GLSNALKK226 pKa = 10.41ALGWNSTLPGSMLVEE241 pKa = 4.83GGALQGRR248 pKa = 11.84GVSPVDD254 pKa = 3.32MDD256 pKa = 4.52AEE258 pKa = 4.38IASRR262 pKa = 11.84TTPDD266 pKa = 3.62LVTDD270 pKa = 3.93MVISSAAEE278 pKa = 3.71MAPHH282 pKa = 6.39IRR284 pKa = 11.84AILALEE290 pKa = 4.45LPSNSGLGDD299 pKa = 3.8LDD301 pKa = 4.28EE302 pKa = 4.9FWSARR307 pKa = 11.84WLWCVNGAQNRR318 pKa = 11.84ASDD321 pKa = 3.61RR322 pKa = 11.84SLNLPSPVRR331 pKa = 11.84GAKK334 pKa = 9.93RR335 pKa = 11.84YY336 pKa = 9.53RR337 pKa = 11.84RR338 pKa = 11.84MAAEE342 pKa = 4.27EE343 pKa = 4.26VSSNPIYY350 pKa = 10.97DD351 pKa = 3.31WDD353 pKa = 3.64GTTNVSASTKK363 pKa = 10.34LEE365 pKa = 4.31PGKK368 pKa = 10.73SRR370 pKa = 11.84AIFACDD376 pKa = 3.03TSSYY380 pKa = 10.84FAFSWILDD388 pKa = 3.42RR389 pKa = 11.84AQRR392 pKa = 11.84DD393 pKa = 3.59WRR395 pKa = 11.84GHH397 pKa = 5.46RR398 pKa = 11.84VLLNPGEE405 pKa = 4.34GGLYY409 pKa = 10.07GVARR413 pKa = 11.84RR414 pKa = 11.84VKK416 pKa = 9.26GAQGRR421 pKa = 11.84GGVNVMLDD429 pKa = 3.4YY430 pKa = 11.58DD431 pKa = 4.74DD432 pKa = 5.89FNSHH436 pKa = 6.63HH437 pKa = 6.74SLGVQQEE444 pKa = 4.29LTRR447 pKa = 11.84QLCQLYY453 pKa = 9.8NAPSWYY459 pKa = 9.58TSVLVDD465 pKa = 3.8SFDD468 pKa = 3.58RR469 pKa = 11.84MYY471 pKa = 9.63ITHH474 pKa = 6.89RR475 pKa = 11.84GSRR478 pKa = 11.84KK479 pKa = 9.8RR480 pKa = 11.84ILGTLMSGHH489 pKa = 7.14RR490 pKa = 11.84GTSFINSVLNAAYY503 pKa = 9.38IRR505 pKa = 11.84AAVGGAFFDD514 pKa = 3.99RR515 pKa = 11.84LVSLHH520 pKa = 6.86AGDD523 pKa = 4.51DD524 pKa = 3.41AYY526 pKa = 10.81MRR528 pKa = 11.84CSTLAEE534 pKa = 4.16AAHH537 pKa = 5.82VLTRR541 pKa = 11.84CARR544 pKa = 11.84FGCRR548 pKa = 11.84MNPTKK553 pKa = 10.53QSIGFRR559 pKa = 11.84HH560 pKa = 6.48AEE562 pKa = 3.93FLRR565 pKa = 11.84LGIGDD570 pKa = 4.47KK571 pKa = 11.0YY572 pKa = 11.29AVGYY576 pKa = 9.11LCRR579 pKa = 11.84SISTLVAGSWVAPDD593 pKa = 4.15PLSPEE598 pKa = 4.88DD599 pKa = 4.28GLTSAITTVRR609 pKa = 11.84SCINRR614 pKa = 11.84GCPSSLSRR622 pKa = 11.84IIARR626 pKa = 11.84TYY628 pKa = 11.11SSIHH632 pKa = 6.34GYY634 pKa = 7.9PLRR637 pKa = 11.84VLDD640 pKa = 4.69ALLSGAACLEE650 pKa = 4.13SGAVYY655 pKa = 8.76NTDD658 pKa = 3.01YY659 pKa = 10.89TLRR662 pKa = 11.84QYY664 pKa = 10.98RR665 pKa = 11.84VVRR668 pKa = 11.84PLPDD672 pKa = 4.84DD673 pKa = 4.61LPLPPQHH680 pKa = 6.92RR681 pKa = 11.84EE682 pKa = 3.68YY683 pKa = 10.67ATRR686 pKa = 11.84EE687 pKa = 3.69YY688 pKa = 10.59LANHH692 pKa = 6.09VSPIEE697 pKa = 3.87ARR699 pKa = 11.84AAQLASADD707 pKa = 3.86LVRR710 pKa = 11.84IMVRR714 pKa = 11.84SSYY717 pKa = 10.95SKK719 pKa = 10.28GVTRR723 pKa = 11.84HH724 pKa = 5.92PSSPTSRR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84PQLVSLPTVKK743 pKa = 10.03ATGFTTAAEE752 pKa = 4.07LAKK755 pKa = 10.26RR756 pKa = 11.84QPPVGKK762 pKa = 9.69LANFPLLRR770 pKa = 11.84LIEE773 pKa = 4.24ARR775 pKa = 11.84LTDD778 pKa = 3.86DD779 pKa = 4.67QISEE783 pKa = 4.35LLSFNGTPPGGLPPRR798 pKa = 11.84IAAFGVEE805 pKa = 4.12GHH807 pKa = 6.53CCNVIGYY814 pKa = 9.1LPYY817 pKa = 10.71SDD819 pKa = 3.98ACSYY823 pKa = 11.09SKK825 pKa = 10.13RR826 pKa = 11.84TTCDD830 pKa = 3.09NIIAGYY836 pKa = 9.3SVYY839 pKa = 11.07SS840 pKa = 3.59
MM1 pKa = 7.13VFVSGRR7 pKa = 11.84VGEE10 pKa = 4.65FGALGSYY17 pKa = 10.26LHH19 pKa = 7.35DD20 pKa = 5.03LLPEE24 pKa = 4.86LGDD27 pKa = 3.62VTEE30 pKa = 5.08SYY32 pKa = 11.86ADD34 pKa = 3.84LEE36 pKa = 4.35LTEE39 pKa = 4.14QLIKK43 pKa = 10.5FATHH47 pKa = 5.6SVSLRR52 pKa = 11.84GVHH55 pKa = 6.25PLGPVAVSLLCVAYY69 pKa = 9.08PVQCDD74 pKa = 3.76FRR76 pKa = 11.84LSDD79 pKa = 3.54LTAVLRR85 pKa = 11.84HH86 pKa = 5.78SFSFLPQRR94 pKa = 11.84APLLEE99 pKa = 3.99LTGLIEE105 pKa = 4.77RR106 pKa = 11.84RR107 pKa = 11.84VTLRR111 pKa = 11.84KK112 pKa = 9.91APATIGALHH121 pKa = 6.17TKK123 pKa = 8.48VARR126 pKa = 11.84LCNSDD131 pKa = 3.11SAARR135 pKa = 11.84SQLWPPKK142 pKa = 8.94RR143 pKa = 11.84HH144 pKa = 5.7AAAGTKK150 pKa = 10.25VNIRR154 pKa = 11.84LAPLAVALSRR164 pKa = 11.84SYY166 pKa = 11.34GPAWLGRR173 pKa = 11.84ALTPLVGCAEE183 pKa = 4.38DD184 pKa = 4.45AVCCALLLATALEE197 pKa = 4.21PHH199 pKa = 6.84FGHH202 pKa = 7.42LGLEE206 pKa = 4.79VAQSMILEE214 pKa = 4.35PNNAKK219 pKa = 10.3GLSNALKK226 pKa = 10.41ALGWNSTLPGSMLVEE241 pKa = 4.83GGALQGRR248 pKa = 11.84GVSPVDD254 pKa = 3.32MDD256 pKa = 4.52AEE258 pKa = 4.38IASRR262 pKa = 11.84TTPDD266 pKa = 3.62LVTDD270 pKa = 3.93MVISSAAEE278 pKa = 3.71MAPHH282 pKa = 6.39IRR284 pKa = 11.84AILALEE290 pKa = 4.45LPSNSGLGDD299 pKa = 3.8LDD301 pKa = 4.28EE302 pKa = 4.9FWSARR307 pKa = 11.84WLWCVNGAQNRR318 pKa = 11.84ASDD321 pKa = 3.61RR322 pKa = 11.84SLNLPSPVRR331 pKa = 11.84GAKK334 pKa = 9.93RR335 pKa = 11.84YY336 pKa = 9.53RR337 pKa = 11.84RR338 pKa = 11.84MAAEE342 pKa = 4.27EE343 pKa = 4.26VSSNPIYY350 pKa = 10.97DD351 pKa = 3.31WDD353 pKa = 3.64GTTNVSASTKK363 pKa = 10.34LEE365 pKa = 4.31PGKK368 pKa = 10.73SRR370 pKa = 11.84AIFACDD376 pKa = 3.03TSSYY380 pKa = 10.84FAFSWILDD388 pKa = 3.42RR389 pKa = 11.84AQRR392 pKa = 11.84DD393 pKa = 3.59WRR395 pKa = 11.84GHH397 pKa = 5.46RR398 pKa = 11.84VLLNPGEE405 pKa = 4.34GGLYY409 pKa = 10.07GVARR413 pKa = 11.84RR414 pKa = 11.84VKK416 pKa = 9.26GAQGRR421 pKa = 11.84GGVNVMLDD429 pKa = 3.4YY430 pKa = 11.58DD431 pKa = 4.74DD432 pKa = 5.89FNSHH436 pKa = 6.63HH437 pKa = 6.74SLGVQQEE444 pKa = 4.29LTRR447 pKa = 11.84QLCQLYY453 pKa = 9.8NAPSWYY459 pKa = 9.58TSVLVDD465 pKa = 3.8SFDD468 pKa = 3.58RR469 pKa = 11.84MYY471 pKa = 9.63ITHH474 pKa = 6.89RR475 pKa = 11.84GSRR478 pKa = 11.84KK479 pKa = 9.8RR480 pKa = 11.84ILGTLMSGHH489 pKa = 7.14RR490 pKa = 11.84GTSFINSVLNAAYY503 pKa = 9.38IRR505 pKa = 11.84AAVGGAFFDD514 pKa = 3.99RR515 pKa = 11.84LVSLHH520 pKa = 6.86AGDD523 pKa = 4.51DD524 pKa = 3.41AYY526 pKa = 10.81MRR528 pKa = 11.84CSTLAEE534 pKa = 4.16AAHH537 pKa = 5.82VLTRR541 pKa = 11.84CARR544 pKa = 11.84FGCRR548 pKa = 11.84MNPTKK553 pKa = 10.53QSIGFRR559 pKa = 11.84HH560 pKa = 6.48AEE562 pKa = 3.93FLRR565 pKa = 11.84LGIGDD570 pKa = 4.47KK571 pKa = 11.0YY572 pKa = 11.29AVGYY576 pKa = 9.11LCRR579 pKa = 11.84SISTLVAGSWVAPDD593 pKa = 4.15PLSPEE598 pKa = 4.88DD599 pKa = 4.28GLTSAITTVRR609 pKa = 11.84SCINRR614 pKa = 11.84GCPSSLSRR622 pKa = 11.84IIARR626 pKa = 11.84TYY628 pKa = 11.11SSIHH632 pKa = 6.34GYY634 pKa = 7.9PLRR637 pKa = 11.84VLDD640 pKa = 4.69ALLSGAACLEE650 pKa = 4.13SGAVYY655 pKa = 8.76NTDD658 pKa = 3.01YY659 pKa = 10.89TLRR662 pKa = 11.84QYY664 pKa = 10.98RR665 pKa = 11.84VVRR668 pKa = 11.84PLPDD672 pKa = 4.84DD673 pKa = 4.61LPLPPQHH680 pKa = 6.92RR681 pKa = 11.84EE682 pKa = 3.68YY683 pKa = 10.67ATRR686 pKa = 11.84EE687 pKa = 3.69YY688 pKa = 10.59LANHH692 pKa = 6.09VSPIEE697 pKa = 3.87ARR699 pKa = 11.84AAQLASADD707 pKa = 3.86LVRR710 pKa = 11.84IMVRR714 pKa = 11.84SSYY717 pKa = 10.95SKK719 pKa = 10.28GVTRR723 pKa = 11.84HH724 pKa = 5.92PSSPTSRR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84PQLVSLPTVKK743 pKa = 10.03ATGFTTAAEE752 pKa = 4.07LAKK755 pKa = 10.26RR756 pKa = 11.84QPPVGKK762 pKa = 9.69LANFPLLRR770 pKa = 11.84LIEE773 pKa = 4.24ARR775 pKa = 11.84LTDD778 pKa = 3.86DD779 pKa = 4.67QISEE783 pKa = 4.35LLSFNGTPPGGLPPRR798 pKa = 11.84IAAFGVEE805 pKa = 4.12GHH807 pKa = 6.53CCNVIGYY814 pKa = 9.1LPYY817 pKa = 10.71SDD819 pKa = 3.98ACSYY823 pKa = 11.09SKK825 pKa = 10.13RR826 pKa = 11.84TTCDD830 pKa = 3.09NIIAGYY836 pKa = 9.3SVYY839 pKa = 11.07SS840 pKa = 3.59
Molecular weight: 91.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1598 |
758 |
840 |
799.0 |
85.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.203 ± 1.088 | 1.564 ± 0.597 |
5.319 ± 0.494 | 3.63 ± 0.132 |
3.379 ± 0.469 | 8.824 ± 0.706 |
2.378 ± 0.176 | 3.88 ± 0.051 |
1.564 ± 0.51 | 9.825 ± 1.607 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.314 ± 0.171 | 3.442 ± 0.253 |
7.259 ± 1.042 | 2.503 ± 0.089 |
7.822 ± 0.286 | 8.073 ± 0.8 |
5.882 ± 0.384 | 6.884 ± 0.072 |
1.252 ± 0.042 | 3.004 ± 0.328 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
