
Elderberry latent virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Pelarspovirus
Average proteome isoelectric point is 8.14
Get precalculated fractions of proteins
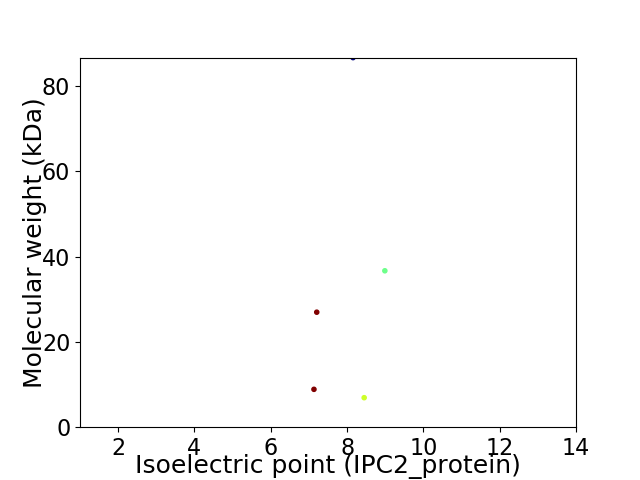
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4MJW8|A0A0B4MJW8_9TOMB Putative movement protein 2 OS=Elderberry latent virus OX=167018 GN=p9 PE=4 SV=1
MM1 pKa = 7.49GLITQCIWFGAYY13 pKa = 9.78VGFKK17 pKa = 8.16VTKK20 pKa = 9.54EE21 pKa = 3.69LALLGPEE28 pKa = 4.25LTSITLSCGFPEE40 pKa = 4.38INPVLAAIGLPTVDD54 pKa = 5.11YY55 pKa = 10.48IPEE58 pKa = 4.08VNQVDD63 pKa = 3.96VLMATNDD70 pKa = 4.53AILHH74 pKa = 5.43TPDD77 pKa = 3.56VEE79 pKa = 5.17LEE81 pKa = 4.26TEE83 pKa = 4.31LEE85 pKa = 4.12DD86 pKa = 5.03SRR88 pKa = 11.84VLTIAEE94 pKa = 4.44PEE96 pKa = 4.2TSRR99 pKa = 11.84VTTVVKK105 pKa = 9.62TRR107 pKa = 11.84RR108 pKa = 11.84PKK110 pKa = 10.02NRR112 pKa = 11.84RR113 pKa = 11.84TFACVLAAEE122 pKa = 4.96AKK124 pKa = 9.3NHH126 pKa = 6.44FGGMPRR132 pKa = 11.84NSRR135 pKa = 11.84ANEE138 pKa = 3.89LSVMKK143 pKa = 10.28YY144 pKa = 10.31LVGKK148 pKa = 9.58CQDD151 pKa = 3.45HH152 pKa = 7.09KK153 pKa = 10.97LTALQTRR160 pKa = 11.84EE161 pKa = 3.6VAAQAFALTFTPDD174 pKa = 3.18ANDD177 pKa = 3.08KK178 pKa = 10.88FIYY181 pKa = 10.05TFLNSEE187 pKa = 4.0DD188 pKa = 4.04AYY190 pKa = 11.03DD191 pKa = 3.75RR192 pKa = 11.84RR193 pKa = 11.84CDD195 pKa = 3.67YY196 pKa = 11.21VKK198 pKa = 10.91SQGVDD203 pKa = 3.6PMWLRR208 pKa = 11.84LLKK211 pKa = 10.59RR212 pKa = 11.84PFSRR216 pKa = 11.84KK217 pKa = 6.31EE218 pKa = 3.55WKK220 pKa = 9.97QVVCKK225 pKa = 10.29FMGFGTRR232 pKa = 11.84EE233 pKa = 3.73AYY235 pKa = 9.43TFIKK239 pKa = 10.56
MM1 pKa = 7.49GLITQCIWFGAYY13 pKa = 9.78VGFKK17 pKa = 8.16VTKK20 pKa = 9.54EE21 pKa = 3.69LALLGPEE28 pKa = 4.25LTSITLSCGFPEE40 pKa = 4.38INPVLAAIGLPTVDD54 pKa = 5.11YY55 pKa = 10.48IPEE58 pKa = 4.08VNQVDD63 pKa = 3.96VLMATNDD70 pKa = 4.53AILHH74 pKa = 5.43TPDD77 pKa = 3.56VEE79 pKa = 5.17LEE81 pKa = 4.26TEE83 pKa = 4.31LEE85 pKa = 4.12DD86 pKa = 5.03SRR88 pKa = 11.84VLTIAEE94 pKa = 4.44PEE96 pKa = 4.2TSRR99 pKa = 11.84VTTVVKK105 pKa = 9.62TRR107 pKa = 11.84RR108 pKa = 11.84PKK110 pKa = 10.02NRR112 pKa = 11.84RR113 pKa = 11.84TFACVLAAEE122 pKa = 4.96AKK124 pKa = 9.3NHH126 pKa = 6.44FGGMPRR132 pKa = 11.84NSRR135 pKa = 11.84ANEE138 pKa = 3.89LSVMKK143 pKa = 10.28YY144 pKa = 10.31LVGKK148 pKa = 9.58CQDD151 pKa = 3.45HH152 pKa = 7.09KK153 pKa = 10.97LTALQTRR160 pKa = 11.84EE161 pKa = 3.6VAAQAFALTFTPDD174 pKa = 3.18ANDD177 pKa = 3.08KK178 pKa = 10.88FIYY181 pKa = 10.05TFLNSEE187 pKa = 4.0DD188 pKa = 4.04AYY190 pKa = 11.03DD191 pKa = 3.75RR192 pKa = 11.84RR193 pKa = 11.84CDD195 pKa = 3.67YY196 pKa = 11.21VKK198 pKa = 10.91SQGVDD203 pKa = 3.6PMWLRR208 pKa = 11.84LLKK211 pKa = 10.59RR212 pKa = 11.84PFSRR216 pKa = 11.84KK217 pKa = 6.31EE218 pKa = 3.55WKK220 pKa = 9.97QVVCKK225 pKa = 10.29FMGFGTRR232 pKa = 11.84EE233 pKa = 3.73AYY235 pKa = 9.43TFIKK239 pKa = 10.56
Molecular weight: 26.98 kDa
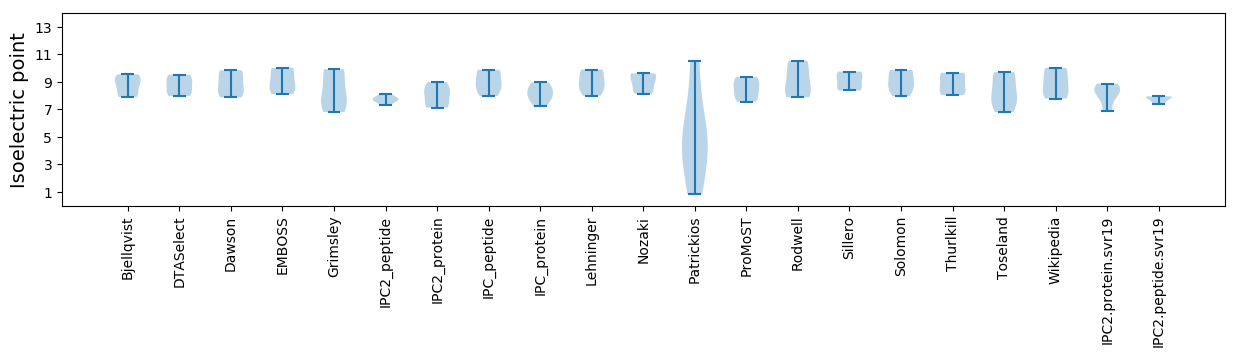
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q911J7|Q911J7_9TOMB Capsid protein OS=Elderberry latent virus OX=167018 GN=p37 PE=3 SV=1
MM1 pKa = 7.53ANKK4 pKa = 10.35SPVVVAAAGRR14 pKa = 11.84GEE16 pKa = 4.08LWAVKK21 pKa = 10.08LSSLGWRR28 pKa = 11.84SLSKK32 pKa = 10.46AQKK35 pKa = 9.96ASARR39 pKa = 11.84AAGIGAVPQSAIIPRR54 pKa = 11.84TTRR57 pKa = 11.84LIAGNPTNSRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84DD71 pKa = 3.4APGNAQKK78 pKa = 11.43SMTVFKK84 pKa = 11.25SEE86 pKa = 4.33FLGEE90 pKa = 3.78LHH92 pKa = 7.27ADD94 pKa = 3.36NTIRR98 pKa = 11.84SYY100 pKa = 11.62KK101 pKa = 10.0LDD103 pKa = 3.58PRR105 pKa = 11.84NVRR108 pKa = 11.84TFPHH112 pKa = 6.46ISDD115 pKa = 4.47LARR118 pKa = 11.84GFNKK122 pKa = 10.41YY123 pKa = 10.29KK124 pKa = 9.74FTDD127 pKa = 3.29VKK129 pKa = 10.68VRR131 pKa = 11.84YY132 pKa = 8.96SSKK135 pKa = 10.68VQDD138 pKa = 4.1SEE140 pKa = 4.7CGLTIAFSADD150 pKa = 3.16SSDD153 pKa = 3.36PAPRR157 pKa = 11.84GKK159 pKa = 10.41FDD161 pKa = 5.19LYY163 pKa = 11.45ALGSKK168 pKa = 10.39VEE170 pKa = 4.36GAAHH174 pKa = 6.93KK175 pKa = 10.46NLLYY179 pKa = 9.92TVPVDD184 pKa = 2.89KK185 pKa = 10.9GQRR188 pKa = 11.84FLRR191 pKa = 11.84DD192 pKa = 3.47GEE194 pKa = 4.16GDD196 pKa = 3.31NAKK199 pKa = 10.26EE200 pKa = 3.57VDD202 pKa = 3.25AGVIYY207 pKa = 11.05VLMDD211 pKa = 4.13GAHH214 pKa = 6.63DD215 pKa = 3.73TRR217 pKa = 11.84AGEE220 pKa = 4.23LFLEE224 pKa = 4.64VTVVLSQPTYY234 pKa = 8.64NQRR237 pKa = 11.84STQYY241 pKa = 10.77ISGTEE246 pKa = 3.82HH247 pKa = 7.08RR248 pKa = 11.84GGPNYY253 pKa = 10.51VNITKK258 pKa = 10.23SANTMTLTFQAAGTFLLCSYY278 pKa = 10.53SSPLEE283 pKa = 3.85KK284 pKa = 10.35VGRR287 pKa = 11.84LSLADD292 pKa = 3.89ADD294 pKa = 3.97QSQVDD299 pKa = 3.94SANNSSNIIEE309 pKa = 4.38AVVTTPGGSIVYY321 pKa = 8.25YY322 pKa = 10.09FKK324 pKa = 11.05SRR326 pKa = 11.84EE327 pKa = 3.93QMSFRR332 pKa = 11.84AYY334 pKa = 9.52VGRR337 pKa = 11.84MM338 pKa = 3.2
MM1 pKa = 7.53ANKK4 pKa = 10.35SPVVVAAAGRR14 pKa = 11.84GEE16 pKa = 4.08LWAVKK21 pKa = 10.08LSSLGWRR28 pKa = 11.84SLSKK32 pKa = 10.46AQKK35 pKa = 9.96ASARR39 pKa = 11.84AAGIGAVPQSAIIPRR54 pKa = 11.84TTRR57 pKa = 11.84LIAGNPTNSRR67 pKa = 11.84RR68 pKa = 11.84TRR70 pKa = 11.84DD71 pKa = 3.4APGNAQKK78 pKa = 11.43SMTVFKK84 pKa = 11.25SEE86 pKa = 4.33FLGEE90 pKa = 3.78LHH92 pKa = 7.27ADD94 pKa = 3.36NTIRR98 pKa = 11.84SYY100 pKa = 11.62KK101 pKa = 10.0LDD103 pKa = 3.58PRR105 pKa = 11.84NVRR108 pKa = 11.84TFPHH112 pKa = 6.46ISDD115 pKa = 4.47LARR118 pKa = 11.84GFNKK122 pKa = 10.41YY123 pKa = 10.29KK124 pKa = 9.74FTDD127 pKa = 3.29VKK129 pKa = 10.68VRR131 pKa = 11.84YY132 pKa = 8.96SSKK135 pKa = 10.68VQDD138 pKa = 4.1SEE140 pKa = 4.7CGLTIAFSADD150 pKa = 3.16SSDD153 pKa = 3.36PAPRR157 pKa = 11.84GKK159 pKa = 10.41FDD161 pKa = 5.19LYY163 pKa = 11.45ALGSKK168 pKa = 10.39VEE170 pKa = 4.36GAAHH174 pKa = 6.93KK175 pKa = 10.46NLLYY179 pKa = 9.92TVPVDD184 pKa = 2.89KK185 pKa = 10.9GQRR188 pKa = 11.84FLRR191 pKa = 11.84DD192 pKa = 3.47GEE194 pKa = 4.16GDD196 pKa = 3.31NAKK199 pKa = 10.26EE200 pKa = 3.57VDD202 pKa = 3.25AGVIYY207 pKa = 11.05VLMDD211 pKa = 4.13GAHH214 pKa = 6.63DD215 pKa = 3.73TRR217 pKa = 11.84AGEE220 pKa = 4.23LFLEE224 pKa = 4.64VTVVLSQPTYY234 pKa = 8.64NQRR237 pKa = 11.84STQYY241 pKa = 10.77ISGTEE246 pKa = 3.82HH247 pKa = 7.08RR248 pKa = 11.84GGPNYY253 pKa = 10.51VNITKK258 pKa = 10.23SANTMTLTFQAAGTFLLCSYY278 pKa = 10.53SSPLEE283 pKa = 3.85KK284 pKa = 10.35VGRR287 pKa = 11.84LSLADD292 pKa = 3.89ADD294 pKa = 3.97QSQVDD299 pKa = 3.94SANNSSNIIEE309 pKa = 4.38AVVTTPGGSIVYY321 pKa = 8.25YY322 pKa = 10.09FKK324 pKa = 11.05SRR326 pKa = 11.84EE327 pKa = 3.93QMSFRR332 pKa = 11.84AYY334 pKa = 9.52VGRR337 pKa = 11.84MM338 pKa = 3.2
Molecular weight: 36.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1485 |
64 |
762 |
297.0 |
33.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.609 ± 1.205 | 1.751 ± 0.507 |
5.32 ± 0.459 | 5.32 ± 0.952 |
4.512 ± 0.267 | 6.801 ± 0.526 |
2.155 ± 0.32 | 4.444 ± 0.229 |
6.128 ± 0.441 | 9.091 ± 0.868 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.359 | 3.838 ± 0.335 |
4.714 ± 0.407 | 3.03 ± 0.253 |
6.667 ± 0.605 | 7.003 ± 1.519 |
6.397 ± 0.752 | 7.879 ± 0.239 |
1.212 ± 0.379 | 3.636 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
