
Botrimarina mediterranea
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales;
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
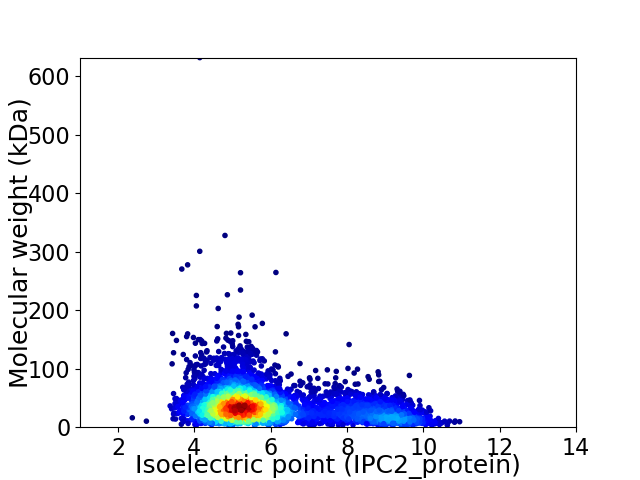
Virtual 2D-PAGE plot for 4587 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
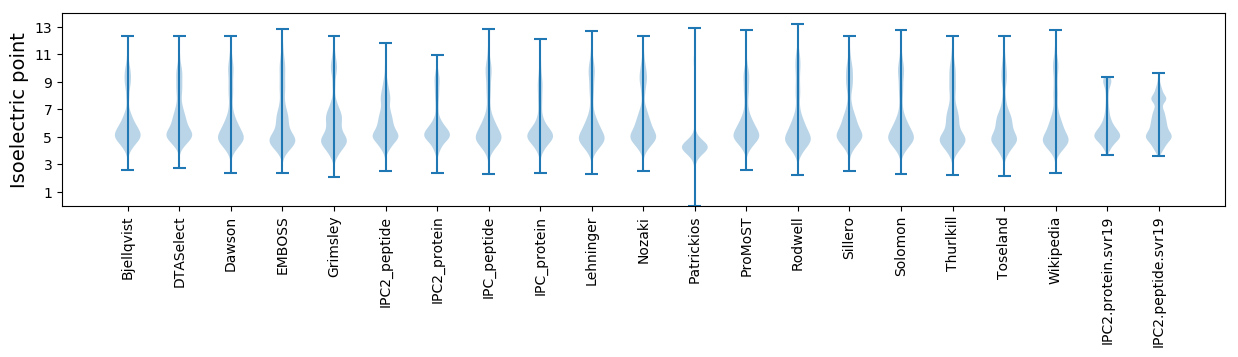
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518KA73|A0A518KA73_9BACT Anhydro-N-acetylmuramic acid kinase OS=Botrimarina mediterranea OX=2528022 GN=anmK_2 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.03FVQHH6 pKa = 6.46AASVAACLAAAVSTPLSVTAQQLSGDD32 pKa = 4.15YY33 pKa = 11.34LMDD36 pKa = 4.91DD37 pKa = 3.89ADD39 pKa = 3.67IAFLGTAYY47 pKa = 10.61LGRR50 pKa = 11.84VGFSVSSAGDD60 pKa = 3.5FNGDD64 pKa = 3.39GFDD67 pKa = 4.96DD68 pKa = 4.47FLFSAPNAPPNSQFSGEE85 pKa = 4.18TFLLYY90 pKa = 10.77GGAEE94 pKa = 4.1TGNGQRR100 pKa = 11.84TLSPMDD106 pKa = 4.15ADD108 pKa = 3.62VHH110 pKa = 5.46FVGIEE115 pKa = 4.06GQHH118 pKa = 5.39AGYY121 pKa = 9.7RR122 pKa = 11.84VSSAGDD128 pKa = 3.61LNLDD132 pKa = 3.88GLDD135 pKa = 4.23DD136 pKa = 4.71LLIATADD143 pKa = 3.54FSEE146 pKa = 4.38RR147 pKa = 11.84NKK149 pKa = 10.37VYY151 pKa = 10.47LVYY154 pKa = 10.69GRR156 pKa = 11.84PDD158 pKa = 3.29ANQLTGEE165 pKa = 4.28FQLSDD170 pKa = 3.24SDD172 pKa = 3.61ATFVLEE178 pKa = 4.33RR179 pKa = 11.84SSDD182 pKa = 3.66PGPVGLTPAGDD193 pKa = 3.51IDD195 pKa = 4.53GDD197 pKa = 4.11GAADD201 pKa = 4.57LLIGDD206 pKa = 3.98YY207 pKa = 10.05TYY209 pKa = 11.56GSGYY213 pKa = 9.5PGRR216 pKa = 11.84AYY218 pKa = 10.86LFYY221 pKa = 10.8GNPDD225 pKa = 3.14ASLSGGIDD233 pKa = 3.06IASADD238 pKa = 3.42ATFDD242 pKa = 3.1VDD244 pKa = 5.75FRR246 pKa = 11.84DD247 pKa = 3.65GTLGYY252 pKa = 9.58SVSSAGDD259 pKa = 3.63LNGDD263 pKa = 3.8GLDD266 pKa = 4.0DD267 pKa = 3.85VAIGALGQGEE277 pKa = 4.6VYY279 pKa = 10.4LYY281 pKa = 10.36YY282 pKa = 10.83GRR284 pKa = 11.84PTSEE288 pKa = 4.32RR289 pKa = 11.84IAGRR293 pKa = 11.84IGRR296 pKa = 11.84GEE298 pKa = 3.9ADD300 pKa = 3.13AVFSVYY306 pKa = 10.56GAGRR310 pKa = 11.84SLSTAGDD317 pKa = 3.9LNGDD321 pKa = 4.0GLNDD325 pKa = 3.85LVIGASDD332 pKa = 3.18MDD334 pKa = 4.23YY335 pKa = 11.34NGEE338 pKa = 4.03EE339 pKa = 3.86SGAAYY344 pKa = 10.73LLYY347 pKa = 10.84GKK349 pKa = 10.33AGDD352 pKa = 3.82NAWRR356 pKa = 11.84GEE358 pKa = 3.8IDD360 pKa = 4.68LDD362 pKa = 3.76NADD365 pKa = 3.37VRR367 pKa = 11.84FEE369 pKa = 4.08GVAEE373 pKa = 4.19GDD375 pKa = 3.29KK376 pKa = 11.06AGYY379 pKa = 8.81QVSSAGDD386 pKa = 3.5VNGDD390 pKa = 4.04GIDD393 pKa = 3.86DD394 pKa = 4.45LLIGAYY400 pKa = 8.56QAQGDD405 pKa = 4.27PNDD408 pKa = 3.99YY409 pKa = 11.44ADD411 pKa = 4.07GQQGGKK417 pKa = 8.3TYY419 pKa = 10.71LVYY422 pKa = 10.64GRR424 pKa = 11.84RR425 pKa = 11.84GGVLDD430 pKa = 3.77EE431 pKa = 5.04AVYY434 pKa = 11.13DD435 pKa = 4.53LDD437 pKa = 3.81NADD440 pKa = 3.64AVFIGGYY447 pKa = 9.49RR448 pKa = 11.84DD449 pKa = 4.47FIGSAVDD456 pKa = 3.35STGDD460 pKa = 3.66LNGDD464 pKa = 3.72GLADD468 pKa = 3.34IVLGAHH474 pKa = 6.94ASDD477 pKa = 3.52TVRR480 pKa = 11.84LSEE483 pKa = 3.84GAVYY487 pKa = 10.27IFYY490 pKa = 10.49GRR492 pKa = 11.84PVPEE496 pKa = 4.19PSAAVLTIAAAAAFSSTFRR515 pKa = 11.84GKK517 pKa = 9.31PVCQFSEE524 pKa = 4.23
MM1 pKa = 7.57KK2 pKa = 10.03FVQHH6 pKa = 6.46AASVAACLAAAVSTPLSVTAQQLSGDD32 pKa = 4.15YY33 pKa = 11.34LMDD36 pKa = 4.91DD37 pKa = 3.89ADD39 pKa = 3.67IAFLGTAYY47 pKa = 10.61LGRR50 pKa = 11.84VGFSVSSAGDD60 pKa = 3.5FNGDD64 pKa = 3.39GFDD67 pKa = 4.96DD68 pKa = 4.47FLFSAPNAPPNSQFSGEE85 pKa = 4.18TFLLYY90 pKa = 10.77GGAEE94 pKa = 4.1TGNGQRR100 pKa = 11.84TLSPMDD106 pKa = 4.15ADD108 pKa = 3.62VHH110 pKa = 5.46FVGIEE115 pKa = 4.06GQHH118 pKa = 5.39AGYY121 pKa = 9.7RR122 pKa = 11.84VSSAGDD128 pKa = 3.61LNLDD132 pKa = 3.88GLDD135 pKa = 4.23DD136 pKa = 4.71LLIATADD143 pKa = 3.54FSEE146 pKa = 4.38RR147 pKa = 11.84NKK149 pKa = 10.37VYY151 pKa = 10.47LVYY154 pKa = 10.69GRR156 pKa = 11.84PDD158 pKa = 3.29ANQLTGEE165 pKa = 4.28FQLSDD170 pKa = 3.24SDD172 pKa = 3.61ATFVLEE178 pKa = 4.33RR179 pKa = 11.84SSDD182 pKa = 3.66PGPVGLTPAGDD193 pKa = 3.51IDD195 pKa = 4.53GDD197 pKa = 4.11GAADD201 pKa = 4.57LLIGDD206 pKa = 3.98YY207 pKa = 10.05TYY209 pKa = 11.56GSGYY213 pKa = 9.5PGRR216 pKa = 11.84AYY218 pKa = 10.86LFYY221 pKa = 10.8GNPDD225 pKa = 3.14ASLSGGIDD233 pKa = 3.06IASADD238 pKa = 3.42ATFDD242 pKa = 3.1VDD244 pKa = 5.75FRR246 pKa = 11.84DD247 pKa = 3.65GTLGYY252 pKa = 9.58SVSSAGDD259 pKa = 3.63LNGDD263 pKa = 3.8GLDD266 pKa = 4.0DD267 pKa = 3.85VAIGALGQGEE277 pKa = 4.6VYY279 pKa = 10.4LYY281 pKa = 10.36YY282 pKa = 10.83GRR284 pKa = 11.84PTSEE288 pKa = 4.32RR289 pKa = 11.84IAGRR293 pKa = 11.84IGRR296 pKa = 11.84GEE298 pKa = 3.9ADD300 pKa = 3.13AVFSVYY306 pKa = 10.56GAGRR310 pKa = 11.84SLSTAGDD317 pKa = 3.9LNGDD321 pKa = 4.0GLNDD325 pKa = 3.85LVIGASDD332 pKa = 3.18MDD334 pKa = 4.23YY335 pKa = 11.34NGEE338 pKa = 4.03EE339 pKa = 3.86SGAAYY344 pKa = 10.73LLYY347 pKa = 10.84GKK349 pKa = 10.33AGDD352 pKa = 3.82NAWRR356 pKa = 11.84GEE358 pKa = 3.8IDD360 pKa = 4.68LDD362 pKa = 3.76NADD365 pKa = 3.37VRR367 pKa = 11.84FEE369 pKa = 4.08GVAEE373 pKa = 4.19GDD375 pKa = 3.29KK376 pKa = 11.06AGYY379 pKa = 8.81QVSSAGDD386 pKa = 3.5VNGDD390 pKa = 4.04GIDD393 pKa = 3.86DD394 pKa = 4.45LLIGAYY400 pKa = 8.56QAQGDD405 pKa = 4.27PNDD408 pKa = 3.99YY409 pKa = 11.44ADD411 pKa = 4.07GQQGGKK417 pKa = 8.3TYY419 pKa = 10.71LVYY422 pKa = 10.64GRR424 pKa = 11.84RR425 pKa = 11.84GGVLDD430 pKa = 3.77EE431 pKa = 5.04AVYY434 pKa = 11.13DD435 pKa = 4.53LDD437 pKa = 3.81NADD440 pKa = 3.64AVFIGGYY447 pKa = 9.49RR448 pKa = 11.84DD449 pKa = 4.47FIGSAVDD456 pKa = 3.35STGDD460 pKa = 3.66LNGDD464 pKa = 3.72GLADD468 pKa = 3.34IVLGAHH474 pKa = 6.94ASDD477 pKa = 3.52TVRR480 pKa = 11.84LSEE483 pKa = 3.84GAVYY487 pKa = 10.27IFYY490 pKa = 10.49GRR492 pKa = 11.84PVPEE496 pKa = 4.19PSAAVLTIAAAAAFSSTFRR515 pKa = 11.84GKK517 pKa = 9.31PVCQFSEE524 pKa = 4.23
Molecular weight: 54.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518KC76|A0A518KC76_9BACT Uncharacterized protein OS=Botrimarina mediterranea OX=2528022 GN=Spa11_36170 PE=4 SV=1
MM1 pKa = 7.31NFSDD5 pKa = 3.99PNLWALLAWLFWCYY19 pKa = 10.77CFLDD23 pKa = 5.45FKK25 pKa = 10.79AHH27 pKa = 6.7RR28 pKa = 11.84YY29 pKa = 9.59DD30 pKa = 5.61RR31 pKa = 11.84IPDD34 pKa = 3.42DD35 pKa = 3.86RR36 pKa = 11.84LRR38 pKa = 11.84VALAEE43 pKa = 4.21HH44 pKa = 6.55EE45 pKa = 4.03AALRR49 pKa = 11.84KK50 pKa = 8.87EE51 pKa = 4.24AAGKK55 pKa = 10.22GDD57 pKa = 3.72PGRR60 pKa = 11.84VNRR63 pKa = 11.84AKK65 pKa = 10.16RR66 pKa = 11.84DD67 pKa = 3.37VVRR70 pKa = 11.84RR71 pKa = 11.84QKK73 pKa = 10.7AASANRR79 pKa = 11.84RR80 pKa = 11.84AHH82 pKa = 5.64QRR84 pKa = 11.84LHH86 pKa = 6.93QEE88 pKa = 3.62VAAANWPVLGRR99 pKa = 11.84VACALVASFLWCHH112 pKa = 6.15ILASVLEE119 pKa = 4.72CARR122 pKa = 11.84QTLPRR127 pKa = 11.84PVAPCVSTRR136 pKa = 11.84PPVSDD141 pKa = 4.69PSTQSTARR149 pKa = 11.84DD150 pKa = 3.38RR151 pKa = 11.84KK152 pKa = 10.54SPALCSSEE160 pKa = 5.09RR161 pKa = 11.84FAQRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84GAPII171 pKa = 3.51
MM1 pKa = 7.31NFSDD5 pKa = 3.99PNLWALLAWLFWCYY19 pKa = 10.77CFLDD23 pKa = 5.45FKK25 pKa = 10.79AHH27 pKa = 6.7RR28 pKa = 11.84YY29 pKa = 9.59DD30 pKa = 5.61RR31 pKa = 11.84IPDD34 pKa = 3.42DD35 pKa = 3.86RR36 pKa = 11.84LRR38 pKa = 11.84VALAEE43 pKa = 4.21HH44 pKa = 6.55EE45 pKa = 4.03AALRR49 pKa = 11.84KK50 pKa = 8.87EE51 pKa = 4.24AAGKK55 pKa = 10.22GDD57 pKa = 3.72PGRR60 pKa = 11.84VNRR63 pKa = 11.84AKK65 pKa = 10.16RR66 pKa = 11.84DD67 pKa = 3.37VVRR70 pKa = 11.84RR71 pKa = 11.84QKK73 pKa = 10.7AASANRR79 pKa = 11.84RR80 pKa = 11.84AHH82 pKa = 5.64QRR84 pKa = 11.84LHH86 pKa = 6.93QEE88 pKa = 3.62VAAANWPVLGRR99 pKa = 11.84VACALVASFLWCHH112 pKa = 6.15ILASVLEE119 pKa = 4.72CARR122 pKa = 11.84QTLPRR127 pKa = 11.84PVAPCVSTRR136 pKa = 11.84PPVSDD141 pKa = 4.69PSTQSTARR149 pKa = 11.84DD150 pKa = 3.38RR151 pKa = 11.84KK152 pKa = 10.54SPALCSSEE160 pKa = 5.09RR161 pKa = 11.84FAQRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84GAPII171 pKa = 3.51
Molecular weight: 19.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1667008 |
29 |
5868 |
363.4 |
39.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.69 ± 0.044 | 1.022 ± 0.016 |
6.157 ± 0.035 | 6.344 ± 0.037 |
3.38 ± 0.022 | 8.489 ± 0.045 |
1.948 ± 0.02 | 4.189 ± 0.024 |
3.052 ± 0.032 | 9.86 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.992 ± 0.017 | 2.822 ± 0.032 |
5.412 ± 0.027 | 3.329 ± 0.022 |
6.898 ± 0.042 | 5.969 ± 0.032 |
5.614 ± 0.035 | 7.8 ± 0.027 |
1.542 ± 0.014 | 2.492 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
