
Capybara microvirus Cap1_SP_104
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.59
Get precalculated fractions of proteins
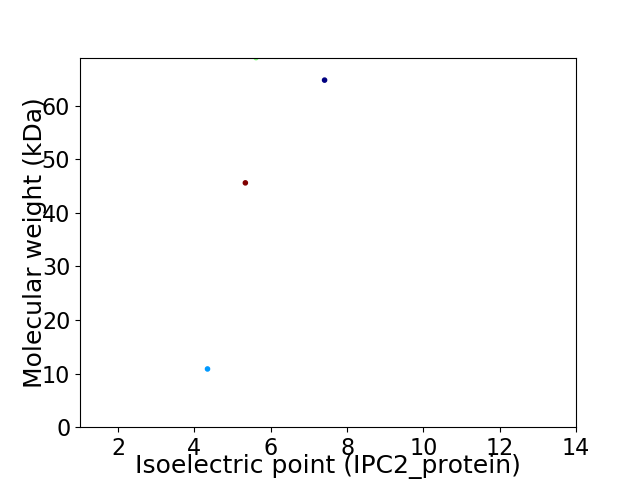
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVN7|A0A4V1FVN7_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_104 OX=2584803 PE=4 SV=1
MM1 pKa = 7.46SVLDD5 pKa = 4.73PIRR8 pKa = 11.84SRR10 pKa = 11.84ISEE13 pKa = 3.94EE14 pKa = 3.53MLYY17 pKa = 10.99DD18 pKa = 4.11GMHH21 pKa = 6.15EE22 pKa = 4.01QLDD25 pKa = 4.0LRR27 pKa = 11.84ILLSADD33 pKa = 3.19GKK35 pKa = 10.12PILTHH40 pKa = 5.58YY41 pKa = 10.49VPSLEE46 pKa = 4.17LSSMYY51 pKa = 10.68DD52 pKa = 3.56LEE54 pKa = 5.27SMLKK58 pKa = 10.52VGITPSGSLHH68 pKa = 6.86CDD70 pKa = 2.9GNSRR74 pKa = 11.84LDD76 pKa = 3.55SFEE79 pKa = 5.59SNITDD84 pKa = 4.16FQTLADD90 pKa = 4.24SFTDD94 pKa = 3.4KK95 pKa = 11.3PEE97 pKa = 3.65
MM1 pKa = 7.46SVLDD5 pKa = 4.73PIRR8 pKa = 11.84SRR10 pKa = 11.84ISEE13 pKa = 3.94EE14 pKa = 3.53MLYY17 pKa = 10.99DD18 pKa = 4.11GMHH21 pKa = 6.15EE22 pKa = 4.01QLDD25 pKa = 4.0LRR27 pKa = 11.84ILLSADD33 pKa = 3.19GKK35 pKa = 10.12PILTHH40 pKa = 5.58YY41 pKa = 10.49VPSLEE46 pKa = 4.17LSSMYY51 pKa = 10.68DD52 pKa = 3.56LEE54 pKa = 5.27SMLKK58 pKa = 10.52VGITPSGSLHH68 pKa = 6.86CDD70 pKa = 2.9GNSRR74 pKa = 11.84LDD76 pKa = 3.55SFEE79 pKa = 5.59SNITDD84 pKa = 4.16FQTLADD90 pKa = 4.24SFTDD94 pKa = 3.4KK95 pKa = 11.3PEE97 pKa = 3.65
Molecular weight: 10.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4W9|A0A4P8W4W9_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_104 OX=2584803 PE=4 SV=1
MM1 pKa = 7.17LTSDD5 pKa = 4.99LIRR8 pKa = 11.84NKK10 pKa = 10.1SCYY13 pKa = 10.11YY14 pKa = 9.26YY15 pKa = 8.12PTRR18 pKa = 11.84NRR20 pKa = 11.84SHH22 pKa = 6.46FRR24 pKa = 11.84VSRR27 pKa = 11.84SDD29 pKa = 3.68LYY31 pKa = 11.07SSRR34 pKa = 11.84VSKK37 pKa = 10.52ILSYY41 pKa = 9.63SVRR44 pKa = 11.84LYY46 pKa = 11.42YY47 pKa = 10.44EE48 pKa = 3.91FLEE51 pKa = 4.49CTEE54 pKa = 3.93CGGRR58 pKa = 11.84VFFYY62 pKa = 10.57TLTYY66 pKa = 10.62NDD68 pKa = 4.14LSIPTLYY75 pKa = 10.81GCPCFDD81 pKa = 4.1YY82 pKa = 11.4NDD84 pKa = 3.25IRR86 pKa = 11.84DD87 pKa = 4.1FVHH90 pKa = 7.13GAFQKK95 pKa = 10.69KK96 pKa = 8.71LCRR99 pKa = 11.84DD100 pKa = 4.7FGCTMKK106 pKa = 10.95YY107 pKa = 10.67AVFCEE112 pKa = 3.9RR113 pKa = 11.84GEE115 pKa = 4.42GKK117 pKa = 10.41GKK119 pKa = 10.45RR120 pKa = 11.84GFGNNPHH127 pKa = 5.7YY128 pKa = 10.68HH129 pKa = 5.9VIFYY133 pKa = 9.23LYY135 pKa = 9.41PKK137 pKa = 9.23EE138 pKa = 3.99GKK140 pKa = 10.29RR141 pKa = 11.84FDD143 pKa = 3.99FVADD147 pKa = 3.71EE148 pKa = 4.26VTLLVRR154 pKa = 11.84KK155 pKa = 9.14YY156 pKa = 8.67WQGDD160 pKa = 3.54CRR162 pKa = 11.84PFDD165 pKa = 3.74VDD167 pKa = 3.86GVNPQSFKK175 pKa = 11.12YY176 pKa = 10.62GIVQVGKK183 pKa = 10.46YY184 pKa = 9.18GAEE187 pKa = 3.86ALDD190 pKa = 3.56YY191 pKa = 10.97RR192 pKa = 11.84AFRR195 pKa = 11.84YY196 pKa = 8.28CAKK199 pKa = 10.6YY200 pKa = 9.96CVKK203 pKa = 10.55DD204 pKa = 3.57AADD207 pKa = 3.65LSVNRR212 pKa = 11.84QIWSSFLKK220 pKa = 10.75DD221 pKa = 3.46FEE223 pKa = 4.5EE224 pKa = 4.76SVKK227 pKa = 10.67NDD229 pKa = 2.96QVLIDD234 pKa = 3.83KK235 pKa = 10.62YY236 pKa = 10.47MYY238 pKa = 11.0AHH240 pKa = 6.14NQYY243 pKa = 10.46SCSIRR248 pKa = 11.84DD249 pKa = 3.56AEE251 pKa = 4.88LSRR254 pKa = 11.84GLPKK258 pKa = 10.26RR259 pKa = 11.84EE260 pKa = 3.53FTFFDD265 pKa = 4.02CATNYY270 pKa = 9.74HH271 pKa = 6.56RR272 pKa = 11.84FFEE275 pKa = 4.23FHH277 pKa = 7.54KK278 pKa = 10.65DD279 pKa = 3.06TLQPILDD286 pKa = 3.93KK287 pKa = 11.02EE288 pKa = 4.11RR289 pKa = 11.84SLFLNRR295 pKa = 11.84FCARR299 pKa = 11.84VRR301 pKa = 11.84ISQGVGISALKK312 pKa = 10.34KK313 pKa = 10.65VIEE316 pKa = 4.85HH317 pKa = 7.59DD318 pKa = 4.26DD319 pKa = 3.86GSLSVPLPSKK329 pKa = 11.0NSFEE333 pKa = 5.48DD334 pKa = 3.36IRR336 pKa = 11.84ISDD339 pKa = 3.65YY340 pKa = 11.34LLRR343 pKa = 11.84KK344 pKa = 9.65LLYY347 pKa = 9.82DD348 pKa = 3.64VKK350 pKa = 10.8KK351 pKa = 11.01DD352 pKa = 3.82PITGNNYY359 pKa = 7.73YY360 pKa = 9.25TLNEE364 pKa = 4.05YY365 pKa = 9.79GQSIKK370 pKa = 10.06MKK372 pKa = 10.01QLQPSIARR380 pKa = 11.84LADD383 pKa = 3.32KK384 pKa = 10.83AFSLVSSNPDD394 pKa = 3.06YY395 pKa = 11.35NSIYY399 pKa = 10.28EE400 pKa = 4.46SYY402 pKa = 11.21SSADD406 pKa = 2.94HH407 pKa = 6.96DD408 pKa = 4.44FRR410 pKa = 11.84WSSGAMLRR418 pKa = 11.84SALSSCTLEE427 pKa = 4.36LCSLYY432 pKa = 11.09ALYY435 pKa = 10.73KK436 pKa = 9.94KK437 pKa = 10.61VYY439 pKa = 8.21EE440 pKa = 4.26FRR442 pKa = 11.84SFAIRR447 pKa = 11.84DD448 pKa = 3.7FQFDD452 pKa = 4.3GVPSLDD458 pKa = 3.88PLADD462 pKa = 3.76YY463 pKa = 11.26YY464 pKa = 11.21STLFDD469 pKa = 3.36FHH471 pKa = 8.01QPILFGQFYY480 pKa = 10.25SHH482 pKa = 7.78DD483 pKa = 3.68GSEE486 pKa = 4.37CFDD489 pKa = 4.58PYY491 pKa = 10.67FIPYY495 pKa = 8.82SAHH498 pKa = 7.2PIFAPHH504 pKa = 6.7FDD506 pKa = 4.09SFCALDD512 pKa = 3.56ILLDD516 pKa = 3.64YY517 pKa = 10.27ATFQDD522 pKa = 3.67QEE524 pKa = 4.37VKK526 pKa = 10.44SRR528 pKa = 11.84RR529 pKa = 11.84DD530 pKa = 3.14AEE532 pKa = 3.75NRR534 pKa = 11.84RR535 pKa = 11.84IKK537 pKa = 10.69NFHH540 pKa = 5.78NNKK543 pKa = 9.38KK544 pKa = 9.23FKK546 pKa = 10.25HH547 pKa = 5.69YY548 pKa = 10.36SLCPFF553 pKa = 4.6
MM1 pKa = 7.17LTSDD5 pKa = 4.99LIRR8 pKa = 11.84NKK10 pKa = 10.1SCYY13 pKa = 10.11YY14 pKa = 9.26YY15 pKa = 8.12PTRR18 pKa = 11.84NRR20 pKa = 11.84SHH22 pKa = 6.46FRR24 pKa = 11.84VSRR27 pKa = 11.84SDD29 pKa = 3.68LYY31 pKa = 11.07SSRR34 pKa = 11.84VSKK37 pKa = 10.52ILSYY41 pKa = 9.63SVRR44 pKa = 11.84LYY46 pKa = 11.42YY47 pKa = 10.44EE48 pKa = 3.91FLEE51 pKa = 4.49CTEE54 pKa = 3.93CGGRR58 pKa = 11.84VFFYY62 pKa = 10.57TLTYY66 pKa = 10.62NDD68 pKa = 4.14LSIPTLYY75 pKa = 10.81GCPCFDD81 pKa = 4.1YY82 pKa = 11.4NDD84 pKa = 3.25IRR86 pKa = 11.84DD87 pKa = 4.1FVHH90 pKa = 7.13GAFQKK95 pKa = 10.69KK96 pKa = 8.71LCRR99 pKa = 11.84DD100 pKa = 4.7FGCTMKK106 pKa = 10.95YY107 pKa = 10.67AVFCEE112 pKa = 3.9RR113 pKa = 11.84GEE115 pKa = 4.42GKK117 pKa = 10.41GKK119 pKa = 10.45RR120 pKa = 11.84GFGNNPHH127 pKa = 5.7YY128 pKa = 10.68HH129 pKa = 5.9VIFYY133 pKa = 9.23LYY135 pKa = 9.41PKK137 pKa = 9.23EE138 pKa = 3.99GKK140 pKa = 10.29RR141 pKa = 11.84FDD143 pKa = 3.99FVADD147 pKa = 3.71EE148 pKa = 4.26VTLLVRR154 pKa = 11.84KK155 pKa = 9.14YY156 pKa = 8.67WQGDD160 pKa = 3.54CRR162 pKa = 11.84PFDD165 pKa = 3.74VDD167 pKa = 3.86GVNPQSFKK175 pKa = 11.12YY176 pKa = 10.62GIVQVGKK183 pKa = 10.46YY184 pKa = 9.18GAEE187 pKa = 3.86ALDD190 pKa = 3.56YY191 pKa = 10.97RR192 pKa = 11.84AFRR195 pKa = 11.84YY196 pKa = 8.28CAKK199 pKa = 10.6YY200 pKa = 9.96CVKK203 pKa = 10.55DD204 pKa = 3.57AADD207 pKa = 3.65LSVNRR212 pKa = 11.84QIWSSFLKK220 pKa = 10.75DD221 pKa = 3.46FEE223 pKa = 4.5EE224 pKa = 4.76SVKK227 pKa = 10.67NDD229 pKa = 2.96QVLIDD234 pKa = 3.83KK235 pKa = 10.62YY236 pKa = 10.47MYY238 pKa = 11.0AHH240 pKa = 6.14NQYY243 pKa = 10.46SCSIRR248 pKa = 11.84DD249 pKa = 3.56AEE251 pKa = 4.88LSRR254 pKa = 11.84GLPKK258 pKa = 10.26RR259 pKa = 11.84EE260 pKa = 3.53FTFFDD265 pKa = 4.02CATNYY270 pKa = 9.74HH271 pKa = 6.56RR272 pKa = 11.84FFEE275 pKa = 4.23FHH277 pKa = 7.54KK278 pKa = 10.65DD279 pKa = 3.06TLQPILDD286 pKa = 3.93KK287 pKa = 11.02EE288 pKa = 4.11RR289 pKa = 11.84SLFLNRR295 pKa = 11.84FCARR299 pKa = 11.84VRR301 pKa = 11.84ISQGVGISALKK312 pKa = 10.34KK313 pKa = 10.65VIEE316 pKa = 4.85HH317 pKa = 7.59DD318 pKa = 4.26DD319 pKa = 3.86GSLSVPLPSKK329 pKa = 11.0NSFEE333 pKa = 5.48DD334 pKa = 3.36IRR336 pKa = 11.84ISDD339 pKa = 3.65YY340 pKa = 11.34LLRR343 pKa = 11.84KK344 pKa = 9.65LLYY347 pKa = 9.82DD348 pKa = 3.64VKK350 pKa = 10.8KK351 pKa = 11.01DD352 pKa = 3.82PITGNNYY359 pKa = 7.73YY360 pKa = 9.25TLNEE364 pKa = 4.05YY365 pKa = 9.79GQSIKK370 pKa = 10.06MKK372 pKa = 10.01QLQPSIARR380 pKa = 11.84LADD383 pKa = 3.32KK384 pKa = 10.83AFSLVSSNPDD394 pKa = 3.06YY395 pKa = 11.35NSIYY399 pKa = 10.28EE400 pKa = 4.46SYY402 pKa = 11.21SSADD406 pKa = 2.94HH407 pKa = 6.96DD408 pKa = 4.44FRR410 pKa = 11.84WSSGAMLRR418 pKa = 11.84SALSSCTLEE427 pKa = 4.36LCSLYY432 pKa = 11.09ALYY435 pKa = 10.73KK436 pKa = 9.94KK437 pKa = 10.61VYY439 pKa = 8.21EE440 pKa = 4.26FRR442 pKa = 11.84SFAIRR447 pKa = 11.84DD448 pKa = 3.7FQFDD452 pKa = 4.3GVPSLDD458 pKa = 3.88PLADD462 pKa = 3.76YY463 pKa = 11.26YY464 pKa = 11.21STLFDD469 pKa = 3.36FHH471 pKa = 8.01QPILFGQFYY480 pKa = 10.25SHH482 pKa = 7.78DD483 pKa = 3.68GSEE486 pKa = 4.37CFDD489 pKa = 4.58PYY491 pKa = 10.67FIPYY495 pKa = 8.82SAHH498 pKa = 7.2PIFAPHH504 pKa = 6.7FDD506 pKa = 4.09SFCALDD512 pKa = 3.56ILLDD516 pKa = 3.64YY517 pKa = 10.27ATFQDD522 pKa = 3.67QEE524 pKa = 4.37VKK526 pKa = 10.44SRR528 pKa = 11.84RR529 pKa = 11.84DD530 pKa = 3.14AEE532 pKa = 3.75NRR534 pKa = 11.84RR535 pKa = 11.84IKK537 pKa = 10.69NFHH540 pKa = 5.78NNKK543 pKa = 9.38KK544 pKa = 9.23FKK546 pKa = 10.25HH547 pKa = 5.69YY548 pKa = 10.36SLCPFF553 pKa = 4.6
Molecular weight: 64.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1666 |
97 |
610 |
416.5 |
47.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.942 ± 1.041 | 1.801 ± 0.673 |
6.723 ± 0.765 | 5.402 ± 0.92 |
6.002 ± 1.242 | 5.402 ± 0.702 |
1.981 ± 0.409 | 5.222 ± 0.409 |
5.582 ± 0.631 | 9.364 ± 0.471 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.281 ± 0.611 | 5.222 ± 0.64 |
4.262 ± 0.267 | 3.842 ± 1.295 |
5.402 ± 0.529 | 10.264 ± 0.463 |
4.262 ± 0.581 | 4.562 ± 0.342 |
0.96 ± 0.251 | 5.522 ± 0.891 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
