
Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) (Yeast) (Torulopsis glabrata)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Nakaseomyces; Nakaseomyces/Candida clade; [Candida] glabrata
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
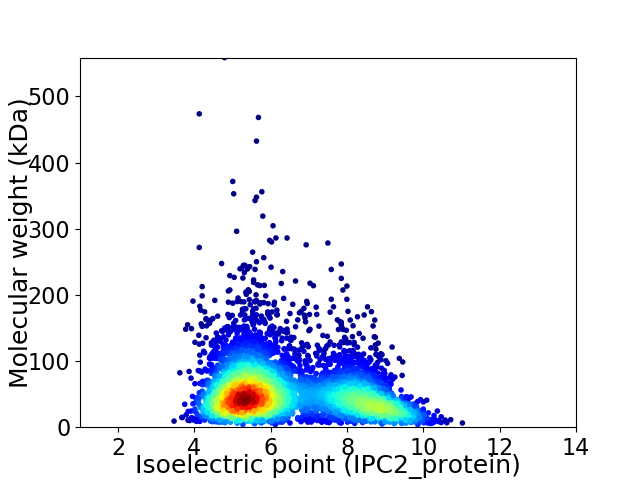
Virtual 2D-PAGE plot for 5201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
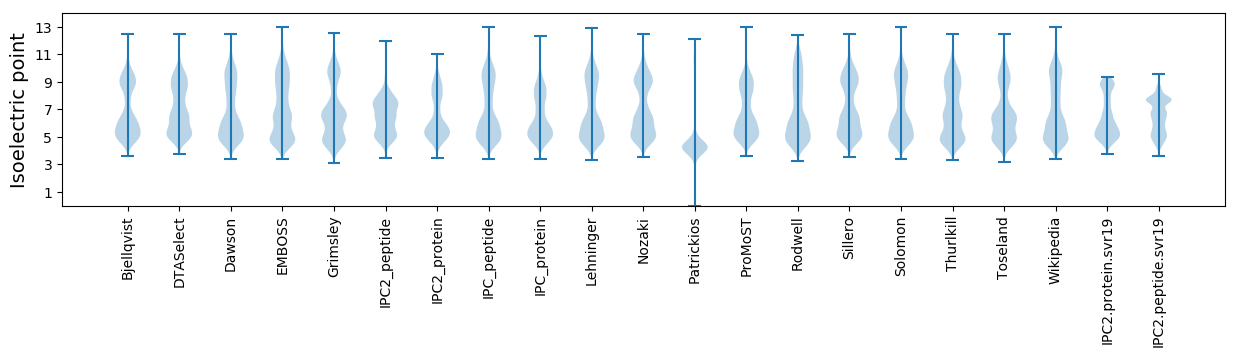
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B4UMX9|B4UMX9_CANGA Uncharacterized protein OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=CAGL0B03899g PE=4 SV=1
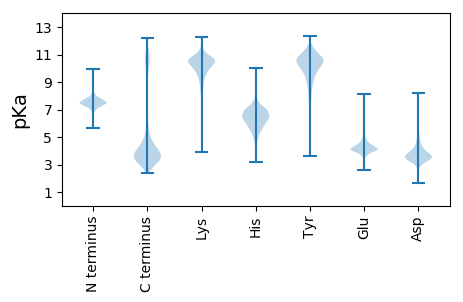
MM1 pKa = 7.21NEE3 pKa = 3.89KK4 pKa = 9.96IFRR7 pKa = 11.84YY8 pKa = 8.33LTLLFLFEE16 pKa = 4.89LASSLPVEE24 pKa = 4.87DD25 pKa = 4.25KK26 pKa = 11.64LNIKK30 pKa = 10.07KK31 pKa = 10.12VFEE34 pKa = 3.96KK35 pKa = 10.44RR36 pKa = 11.84AVDD39 pKa = 4.07FTPFAHH45 pKa = 6.29YY46 pKa = 7.94PRR48 pKa = 11.84PEE50 pKa = 4.59GCSSPPNSVSVGLHH64 pKa = 4.79MNLYY68 pKa = 9.29NYY70 pKa = 9.15PYY72 pKa = 10.41LYY74 pKa = 10.59VKK76 pKa = 10.45PSTKK80 pKa = 10.62GMFTNDD86 pKa = 3.39TNSDD90 pKa = 3.4SDD92 pKa = 4.18GEE94 pKa = 4.35TDD96 pKa = 3.48GDD98 pKa = 3.97SAGGIEE104 pKa = 5.14GRR106 pKa = 11.84AGQCWNPEE114 pKa = 4.0YY115 pKa = 10.27QDD117 pKa = 3.51PNFPRR122 pKa = 11.84FGYY125 pKa = 9.27KK126 pKa = 9.71QYY128 pKa = 11.68GEE130 pKa = 5.0FGASDD135 pKa = 3.89NVNGDD140 pKa = 3.14ISWDD144 pKa = 3.49HH145 pKa = 6.78NEE147 pKa = 3.87FTEE150 pKa = 4.46GCKK153 pKa = 9.77PVLAALPPGYY163 pKa = 10.12NYY165 pKa = 9.85PDD167 pKa = 4.87EE168 pKa = 4.31ITFSNFTMVLSGYY181 pKa = 8.69FKK183 pKa = 10.65PKK185 pKa = 9.3TSGVYY190 pKa = 10.11KK191 pKa = 10.73FEE193 pKa = 4.77LKK195 pKa = 10.66ADD197 pKa = 3.83DD198 pKa = 5.31FILFNFGAKK207 pKa = 9.43NAFEE211 pKa = 4.31CCNRR215 pKa = 11.84EE216 pKa = 3.63EE217 pKa = 4.92SIDD220 pKa = 3.65NFGPYY225 pKa = 8.59VAYY228 pKa = 10.53AMWPNQADD236 pKa = 3.62QEE238 pKa = 4.58LEE240 pKa = 4.13VYY242 pKa = 10.5LFEE245 pKa = 6.18DD246 pKa = 3.4SYY248 pKa = 11.61YY249 pKa = 10.22PLRR252 pKa = 11.84LFYY255 pKa = 11.07NNRR258 pKa = 11.84DD259 pKa = 3.39YY260 pKa = 11.25HH261 pKa = 7.21SKK263 pKa = 10.61FKK265 pKa = 10.68IGFYY269 pKa = 10.53PPGQTTITFDD279 pKa = 3.23FDD281 pKa = 3.85GYY283 pKa = 11.54LFMLDD288 pKa = 3.63DD289 pKa = 4.39TGNEE293 pKa = 3.98CRR295 pKa = 11.84DD296 pKa = 3.67SIRR299 pKa = 11.84YY300 pKa = 7.46RR301 pKa = 11.84TVCDD305 pKa = 4.11DD306 pKa = 3.93KK307 pKa = 11.87VVDD310 pKa = 3.77DD311 pKa = 3.93TTYY314 pKa = 8.5STRR317 pKa = 11.84YY318 pKa = 6.78STSMVPGGGSDD329 pKa = 3.72VITIYY334 pKa = 10.14YY335 pKa = 9.9IRR337 pKa = 11.84MKK339 pKa = 10.74CEE341 pKa = 3.83EE342 pKa = 4.58EE343 pKa = 4.5DD344 pKa = 5.7CLGQWDD350 pKa = 5.16PINNVCYY357 pKa = 10.83SPDD360 pKa = 3.42DD361 pKa = 4.01CADD364 pKa = 4.21DD365 pKa = 3.77GGYY368 pKa = 10.18WNGQMCDD375 pKa = 3.46QSCKK379 pKa = 10.61LEE381 pKa = 4.18GGIVNPDD388 pKa = 3.07TGDD391 pKa = 3.55CDD393 pKa = 3.79KK394 pKa = 11.66SCIEE398 pKa = 4.18SGGFLDD404 pKa = 5.56EE405 pKa = 5.62NGNCDD410 pKa = 3.45TTCRR414 pKa = 11.84DD415 pKa = 3.55DD416 pKa = 5.09GGMLVEE422 pKa = 4.61GQCDD426 pKa = 4.04YY427 pKa = 11.14QCKK430 pKa = 8.82EE431 pKa = 3.7AGGILVGDD439 pKa = 4.3HH440 pKa = 7.42CDD442 pKa = 3.6TTCVDD447 pKa = 3.85SGGKK451 pKa = 10.02LNEE454 pKa = 5.64DD455 pKa = 3.63GTCDD459 pKa = 4.13HH460 pKa = 7.05SCRR463 pKa = 11.84DD464 pKa = 3.12QGGQLDD470 pKa = 4.03EE471 pKa = 5.47SGEE474 pKa = 4.34CDD476 pKa = 3.59TSCKK480 pKa = 10.4DD481 pKa = 3.23SGGMLIEE488 pKa = 4.78GEE490 pKa = 4.73CDD492 pKa = 3.32TSCKK496 pKa = 10.63DD497 pKa = 3.25EE498 pKa = 4.82GGQLDD503 pKa = 4.55EE504 pKa = 5.04NNEE507 pKa = 4.5CINHH511 pKa = 6.33CKK513 pKa = 10.26DD514 pKa = 2.84QGGIIDD520 pKa = 4.27EE521 pKa = 4.89NGNCDD526 pKa = 3.84TSCKK530 pKa = 10.42DD531 pKa = 3.15SGGKK535 pKa = 9.68LDD537 pKa = 4.85EE538 pKa = 5.33NGACDD543 pKa = 3.66TSCRR547 pKa = 11.84DD548 pKa = 3.01EE549 pKa = 4.97GGIIDD554 pKa = 4.91EE555 pKa = 4.95NGNCNTTCRR564 pKa = 11.84DD565 pKa = 3.38DD566 pKa = 4.5GGILVGDD573 pKa = 4.29HH574 pKa = 7.4CDD576 pKa = 3.43TSCRR580 pKa = 11.84DD581 pKa = 3.19AGGILEE587 pKa = 5.57DD588 pKa = 5.02GICNSTCRR596 pKa = 11.84DD597 pKa = 3.26SGGKK601 pKa = 10.36LNDD604 pKa = 5.33DD605 pKa = 3.68GTCDD609 pKa = 3.67YY610 pKa = 11.26SCRR613 pKa = 11.84DD614 pKa = 2.96QGGQLDD620 pKa = 4.13EE621 pKa = 5.96NGDD624 pKa = 3.82CDD626 pKa = 4.25YY627 pKa = 11.37SCKK630 pKa = 10.69NGGGKK635 pKa = 9.67LDD637 pKa = 3.75EE638 pKa = 6.11DD639 pKa = 5.1GNCDD643 pKa = 3.5MSCKK647 pKa = 10.31EE648 pKa = 4.33NGGQLDD654 pKa = 3.85EE655 pKa = 7.07DD656 pKa = 4.76GDD658 pKa = 4.44CDD660 pKa = 5.28LSCKK664 pKa = 10.21EE665 pKa = 5.12DD666 pKa = 3.39GGKK669 pKa = 10.22LDD671 pKa = 4.6GNGDD675 pKa = 3.67CDD677 pKa = 4.07YY678 pKa = 11.24SCKK681 pKa = 10.51EE682 pKa = 3.66EE683 pKa = 4.3GGIIDD688 pKa = 4.6EE689 pKa = 4.92SGNCNTTCKK698 pKa = 10.55DD699 pKa = 3.06SGGQIIDD706 pKa = 3.76GEE708 pKa = 4.54CNYY711 pKa = 10.91GCIEE715 pKa = 4.18VGGVVIDD722 pKa = 4.55GEE724 pKa = 4.7CDD726 pKa = 3.43TSCVDD731 pKa = 4.21SGGILDD737 pKa = 5.01GDD739 pKa = 4.47GNCDD743 pKa = 3.22RR744 pKa = 11.84SCIEE748 pKa = 3.85SGGVLEE754 pKa = 6.27DD755 pKa = 4.18GACNTTCRR763 pKa = 11.84DD764 pKa = 3.03SGGKK768 pKa = 10.32LNVDD772 pKa = 4.18GTCDD776 pKa = 3.38TSCRR780 pKa = 11.84DD781 pKa = 3.54DD782 pKa = 4.35GGMLVEE788 pKa = 4.61GQCDD792 pKa = 4.04YY793 pKa = 11.14QCKK796 pKa = 8.86EE797 pKa = 3.77AGGILEE803 pKa = 5.3DD804 pKa = 4.58GEE806 pKa = 5.47CVFPPTSSSTSEE818 pKa = 4.07TSSTTDD824 pKa = 3.08QSSSQSDD831 pKa = 3.59DD832 pKa = 3.55SSVEE836 pKa = 4.18SSSTPMDD843 pKa = 3.31SSTIEE848 pKa = 4.05SSSSEE853 pKa = 4.05IITSEE858 pKa = 4.19SEE860 pKa = 4.28SEE862 pKa = 4.38SVPIASSSSEE872 pKa = 3.83VMTSEE877 pKa = 4.71SEE879 pKa = 4.33SEE881 pKa = 4.4SIPILTSSSEE891 pKa = 3.85IPIASSSSEE900 pKa = 3.74IIIPEE905 pKa = 4.16SEE907 pKa = 4.3SEE909 pKa = 4.46SVPVASSSSEE919 pKa = 3.71IPVASSSSEE928 pKa = 3.72IIIPEE933 pKa = 4.16SEE935 pKa = 4.34SEE937 pKa = 4.44SVPIASSSSEE947 pKa = 3.77IPVASSSSEE956 pKa = 3.72IIIPEE961 pKa = 4.18SEE963 pKa = 4.29SEE965 pKa = 4.35SIPIEE970 pKa = 4.36STSSEE975 pKa = 4.1AVSSRR980 pKa = 11.84SEE982 pKa = 4.0SDD984 pKa = 2.79SSYY987 pKa = 9.35PTSTPTTTTSTTWPNPIQPTTSNSSPIVSSSTEE1020 pKa = 3.83TPGIAEE1026 pKa = 4.02SSSRR1030 pKa = 11.84ISDD1033 pKa = 3.81LEE1035 pKa = 3.9SSRR1038 pKa = 11.84RR1039 pKa = 11.84TSILGDD1045 pKa = 3.14DD1046 pKa = 3.92RR1047 pKa = 11.84TRR1049 pKa = 11.84TRR1051 pKa = 11.84TTITSTITEE1060 pKa = 4.73DD1061 pKa = 3.52GQIITITTTVTGFVPEE1077 pKa = 4.24PTGEE1081 pKa = 4.24TEE1083 pKa = 4.82GNGSDD1088 pKa = 5.49DD1089 pKa = 4.14EE1090 pKa = 4.46NLEE1093 pKa = 4.1NPIEE1097 pKa = 4.34NNTDD1101 pKa = 2.97EE1102 pKa = 5.07NEE1104 pKa = 4.2FVDD1107 pKa = 5.76PEE1109 pKa = 4.24FTDD1112 pKa = 6.26DD1113 pKa = 5.61IPDD1116 pKa = 4.17DD1117 pKa = 3.94PAGPEE1122 pKa = 4.07DD1123 pKa = 4.06NGNGPVNPEE1132 pKa = 3.76QGNDD1136 pKa = 3.34NDD1138 pKa = 4.42PVNLEE1143 pKa = 3.96DD1144 pKa = 4.28SGNNGAVNPEE1154 pKa = 3.77QAPVNDD1160 pKa = 4.36EE1161 pKa = 4.64DD1162 pKa = 4.9NGGNDD1167 pKa = 3.95PVNPEE1172 pKa = 3.53QDD1174 pKa = 3.86NNNGPANPGQDD1185 pKa = 3.3NTNGSSNSDD1194 pKa = 3.35QIDD1197 pKa = 3.89NNPPTPEE1204 pKa = 3.88EE1205 pKa = 3.83NAGDD1209 pKa = 4.05GNSNGNDD1216 pKa = 3.42PADD1219 pKa = 3.84PSRR1222 pKa = 11.84RR1223 pKa = 11.84TSILGDD1229 pKa = 3.2EE1230 pKa = 4.64RR1231 pKa = 11.84TRR1233 pKa = 11.84ASPGGTSQPTGANGNVNGNSPGANVNGGGTGNSNNNNNNNNNNNNNNNNGNAPNGGDD1290 pKa = 4.24GSNGDD1295 pKa = 3.97ANQGPSRR1302 pKa = 11.84RR1303 pKa = 11.84TSLLGDD1309 pKa = 3.27EE1310 pKa = 4.53RR1311 pKa = 11.84TRR1313 pKa = 11.84AGQAAPTGGSSPNNGVAGDD1332 pKa = 3.66TSNNGGAIVSQQVDD1346 pKa = 3.47DD1347 pKa = 5.02GGAGSPRR1354 pKa = 11.84RR1355 pKa = 11.84TSILGDD1361 pKa = 3.18VRR1363 pKa = 11.84SRR1365 pKa = 11.84TTLNGEE1371 pKa = 4.05VLNFEE1376 pKa = 4.37GSGVRR1381 pKa = 11.84RR1382 pKa = 11.84AVAGNLALPLMMVGLVFNLL1401 pKa = 3.56
MM1 pKa = 7.21NEE3 pKa = 3.89KK4 pKa = 9.96IFRR7 pKa = 11.84YY8 pKa = 8.33LTLLFLFEE16 pKa = 4.89LASSLPVEE24 pKa = 4.87DD25 pKa = 4.25KK26 pKa = 11.64LNIKK30 pKa = 10.07KK31 pKa = 10.12VFEE34 pKa = 3.96KK35 pKa = 10.44RR36 pKa = 11.84AVDD39 pKa = 4.07FTPFAHH45 pKa = 6.29YY46 pKa = 7.94PRR48 pKa = 11.84PEE50 pKa = 4.59GCSSPPNSVSVGLHH64 pKa = 4.79MNLYY68 pKa = 9.29NYY70 pKa = 9.15PYY72 pKa = 10.41LYY74 pKa = 10.59VKK76 pKa = 10.45PSTKK80 pKa = 10.62GMFTNDD86 pKa = 3.39TNSDD90 pKa = 3.4SDD92 pKa = 4.18GEE94 pKa = 4.35TDD96 pKa = 3.48GDD98 pKa = 3.97SAGGIEE104 pKa = 5.14GRR106 pKa = 11.84AGQCWNPEE114 pKa = 4.0YY115 pKa = 10.27QDD117 pKa = 3.51PNFPRR122 pKa = 11.84FGYY125 pKa = 9.27KK126 pKa = 9.71QYY128 pKa = 11.68GEE130 pKa = 5.0FGASDD135 pKa = 3.89NVNGDD140 pKa = 3.14ISWDD144 pKa = 3.49HH145 pKa = 6.78NEE147 pKa = 3.87FTEE150 pKa = 4.46GCKK153 pKa = 9.77PVLAALPPGYY163 pKa = 10.12NYY165 pKa = 9.85PDD167 pKa = 4.87EE168 pKa = 4.31ITFSNFTMVLSGYY181 pKa = 8.69FKK183 pKa = 10.65PKK185 pKa = 9.3TSGVYY190 pKa = 10.11KK191 pKa = 10.73FEE193 pKa = 4.77LKK195 pKa = 10.66ADD197 pKa = 3.83DD198 pKa = 5.31FILFNFGAKK207 pKa = 9.43NAFEE211 pKa = 4.31CCNRR215 pKa = 11.84EE216 pKa = 3.63EE217 pKa = 4.92SIDD220 pKa = 3.65NFGPYY225 pKa = 8.59VAYY228 pKa = 10.53AMWPNQADD236 pKa = 3.62QEE238 pKa = 4.58LEE240 pKa = 4.13VYY242 pKa = 10.5LFEE245 pKa = 6.18DD246 pKa = 3.4SYY248 pKa = 11.61YY249 pKa = 10.22PLRR252 pKa = 11.84LFYY255 pKa = 11.07NNRR258 pKa = 11.84DD259 pKa = 3.39YY260 pKa = 11.25HH261 pKa = 7.21SKK263 pKa = 10.61FKK265 pKa = 10.68IGFYY269 pKa = 10.53PPGQTTITFDD279 pKa = 3.23FDD281 pKa = 3.85GYY283 pKa = 11.54LFMLDD288 pKa = 3.63DD289 pKa = 4.39TGNEE293 pKa = 3.98CRR295 pKa = 11.84DD296 pKa = 3.67SIRR299 pKa = 11.84YY300 pKa = 7.46RR301 pKa = 11.84TVCDD305 pKa = 4.11DD306 pKa = 3.93KK307 pKa = 11.87VVDD310 pKa = 3.77DD311 pKa = 3.93TTYY314 pKa = 8.5STRR317 pKa = 11.84YY318 pKa = 6.78STSMVPGGGSDD329 pKa = 3.72VITIYY334 pKa = 10.14YY335 pKa = 9.9IRR337 pKa = 11.84MKK339 pKa = 10.74CEE341 pKa = 3.83EE342 pKa = 4.58EE343 pKa = 4.5DD344 pKa = 5.7CLGQWDD350 pKa = 5.16PINNVCYY357 pKa = 10.83SPDD360 pKa = 3.42DD361 pKa = 4.01CADD364 pKa = 4.21DD365 pKa = 3.77GGYY368 pKa = 10.18WNGQMCDD375 pKa = 3.46QSCKK379 pKa = 10.61LEE381 pKa = 4.18GGIVNPDD388 pKa = 3.07TGDD391 pKa = 3.55CDD393 pKa = 3.79KK394 pKa = 11.66SCIEE398 pKa = 4.18SGGFLDD404 pKa = 5.56EE405 pKa = 5.62NGNCDD410 pKa = 3.45TTCRR414 pKa = 11.84DD415 pKa = 3.55DD416 pKa = 5.09GGMLVEE422 pKa = 4.61GQCDD426 pKa = 4.04YY427 pKa = 11.14QCKK430 pKa = 8.82EE431 pKa = 3.7AGGILVGDD439 pKa = 4.3HH440 pKa = 7.42CDD442 pKa = 3.6TTCVDD447 pKa = 3.85SGGKK451 pKa = 10.02LNEE454 pKa = 5.64DD455 pKa = 3.63GTCDD459 pKa = 4.13HH460 pKa = 7.05SCRR463 pKa = 11.84DD464 pKa = 3.12QGGQLDD470 pKa = 4.03EE471 pKa = 5.47SGEE474 pKa = 4.34CDD476 pKa = 3.59TSCKK480 pKa = 10.4DD481 pKa = 3.23SGGMLIEE488 pKa = 4.78GEE490 pKa = 4.73CDD492 pKa = 3.32TSCKK496 pKa = 10.63DD497 pKa = 3.25EE498 pKa = 4.82GGQLDD503 pKa = 4.55EE504 pKa = 5.04NNEE507 pKa = 4.5CINHH511 pKa = 6.33CKK513 pKa = 10.26DD514 pKa = 2.84QGGIIDD520 pKa = 4.27EE521 pKa = 4.89NGNCDD526 pKa = 3.84TSCKK530 pKa = 10.42DD531 pKa = 3.15SGGKK535 pKa = 9.68LDD537 pKa = 4.85EE538 pKa = 5.33NGACDD543 pKa = 3.66TSCRR547 pKa = 11.84DD548 pKa = 3.01EE549 pKa = 4.97GGIIDD554 pKa = 4.91EE555 pKa = 4.95NGNCNTTCRR564 pKa = 11.84DD565 pKa = 3.38DD566 pKa = 4.5GGILVGDD573 pKa = 4.29HH574 pKa = 7.4CDD576 pKa = 3.43TSCRR580 pKa = 11.84DD581 pKa = 3.19AGGILEE587 pKa = 5.57DD588 pKa = 5.02GICNSTCRR596 pKa = 11.84DD597 pKa = 3.26SGGKK601 pKa = 10.36LNDD604 pKa = 5.33DD605 pKa = 3.68GTCDD609 pKa = 3.67YY610 pKa = 11.26SCRR613 pKa = 11.84DD614 pKa = 2.96QGGQLDD620 pKa = 4.13EE621 pKa = 5.96NGDD624 pKa = 3.82CDD626 pKa = 4.25YY627 pKa = 11.37SCKK630 pKa = 10.69NGGGKK635 pKa = 9.67LDD637 pKa = 3.75EE638 pKa = 6.11DD639 pKa = 5.1GNCDD643 pKa = 3.5MSCKK647 pKa = 10.31EE648 pKa = 4.33NGGQLDD654 pKa = 3.85EE655 pKa = 7.07DD656 pKa = 4.76GDD658 pKa = 4.44CDD660 pKa = 5.28LSCKK664 pKa = 10.21EE665 pKa = 5.12DD666 pKa = 3.39GGKK669 pKa = 10.22LDD671 pKa = 4.6GNGDD675 pKa = 3.67CDD677 pKa = 4.07YY678 pKa = 11.24SCKK681 pKa = 10.51EE682 pKa = 3.66EE683 pKa = 4.3GGIIDD688 pKa = 4.6EE689 pKa = 4.92SGNCNTTCKK698 pKa = 10.55DD699 pKa = 3.06SGGQIIDD706 pKa = 3.76GEE708 pKa = 4.54CNYY711 pKa = 10.91GCIEE715 pKa = 4.18VGGVVIDD722 pKa = 4.55GEE724 pKa = 4.7CDD726 pKa = 3.43TSCVDD731 pKa = 4.21SGGILDD737 pKa = 5.01GDD739 pKa = 4.47GNCDD743 pKa = 3.22RR744 pKa = 11.84SCIEE748 pKa = 3.85SGGVLEE754 pKa = 6.27DD755 pKa = 4.18GACNTTCRR763 pKa = 11.84DD764 pKa = 3.03SGGKK768 pKa = 10.32LNVDD772 pKa = 4.18GTCDD776 pKa = 3.38TSCRR780 pKa = 11.84DD781 pKa = 3.54DD782 pKa = 4.35GGMLVEE788 pKa = 4.61GQCDD792 pKa = 4.04YY793 pKa = 11.14QCKK796 pKa = 8.86EE797 pKa = 3.77AGGILEE803 pKa = 5.3DD804 pKa = 4.58GEE806 pKa = 5.47CVFPPTSSSTSEE818 pKa = 4.07TSSTTDD824 pKa = 3.08QSSSQSDD831 pKa = 3.59DD832 pKa = 3.55SSVEE836 pKa = 4.18SSSTPMDD843 pKa = 3.31SSTIEE848 pKa = 4.05SSSSEE853 pKa = 4.05IITSEE858 pKa = 4.19SEE860 pKa = 4.28SEE862 pKa = 4.38SVPIASSSSEE872 pKa = 3.83VMTSEE877 pKa = 4.71SEE879 pKa = 4.33SEE881 pKa = 4.4SIPILTSSSEE891 pKa = 3.85IPIASSSSEE900 pKa = 3.74IIIPEE905 pKa = 4.16SEE907 pKa = 4.3SEE909 pKa = 4.46SVPVASSSSEE919 pKa = 3.71IPVASSSSEE928 pKa = 3.72IIIPEE933 pKa = 4.16SEE935 pKa = 4.34SEE937 pKa = 4.44SVPIASSSSEE947 pKa = 3.77IPVASSSSEE956 pKa = 3.72IIIPEE961 pKa = 4.18SEE963 pKa = 4.29SEE965 pKa = 4.35SIPIEE970 pKa = 4.36STSSEE975 pKa = 4.1AVSSRR980 pKa = 11.84SEE982 pKa = 4.0SDD984 pKa = 2.79SSYY987 pKa = 9.35PTSTPTTTTSTTWPNPIQPTTSNSSPIVSSSTEE1020 pKa = 3.83TPGIAEE1026 pKa = 4.02SSSRR1030 pKa = 11.84ISDD1033 pKa = 3.81LEE1035 pKa = 3.9SSRR1038 pKa = 11.84RR1039 pKa = 11.84TSILGDD1045 pKa = 3.14DD1046 pKa = 3.92RR1047 pKa = 11.84TRR1049 pKa = 11.84TRR1051 pKa = 11.84TTITSTITEE1060 pKa = 4.73DD1061 pKa = 3.52GQIITITTTVTGFVPEE1077 pKa = 4.24PTGEE1081 pKa = 4.24TEE1083 pKa = 4.82GNGSDD1088 pKa = 5.49DD1089 pKa = 4.14EE1090 pKa = 4.46NLEE1093 pKa = 4.1NPIEE1097 pKa = 4.34NNTDD1101 pKa = 2.97EE1102 pKa = 5.07NEE1104 pKa = 4.2FVDD1107 pKa = 5.76PEE1109 pKa = 4.24FTDD1112 pKa = 6.26DD1113 pKa = 5.61IPDD1116 pKa = 4.17DD1117 pKa = 3.94PAGPEE1122 pKa = 4.07DD1123 pKa = 4.06NGNGPVNPEE1132 pKa = 3.76QGNDD1136 pKa = 3.34NDD1138 pKa = 4.42PVNLEE1143 pKa = 3.96DD1144 pKa = 4.28SGNNGAVNPEE1154 pKa = 3.77QAPVNDD1160 pKa = 4.36EE1161 pKa = 4.64DD1162 pKa = 4.9NGGNDD1167 pKa = 3.95PVNPEE1172 pKa = 3.53QDD1174 pKa = 3.86NNNGPANPGQDD1185 pKa = 3.3NTNGSSNSDD1194 pKa = 3.35QIDD1197 pKa = 3.89NNPPTPEE1204 pKa = 3.88EE1205 pKa = 3.83NAGDD1209 pKa = 4.05GNSNGNDD1216 pKa = 3.42PADD1219 pKa = 3.84PSRR1222 pKa = 11.84RR1223 pKa = 11.84TSILGDD1229 pKa = 3.2EE1230 pKa = 4.64RR1231 pKa = 11.84TRR1233 pKa = 11.84ASPGGTSQPTGANGNVNGNSPGANVNGGGTGNSNNNNNNNNNNNNNNNNGNAPNGGDD1290 pKa = 4.24GSNGDD1295 pKa = 3.97ANQGPSRR1302 pKa = 11.84RR1303 pKa = 11.84TSLLGDD1309 pKa = 3.27EE1310 pKa = 4.53RR1311 pKa = 11.84TRR1313 pKa = 11.84AGQAAPTGGSSPNNGVAGDD1332 pKa = 3.66TSNNGGAIVSQQVDD1346 pKa = 3.47DD1347 pKa = 5.02GGAGSPRR1354 pKa = 11.84RR1355 pKa = 11.84TSILGDD1361 pKa = 3.18VRR1363 pKa = 11.84SRR1365 pKa = 11.84TTLNGEE1371 pKa = 4.05VLNFEE1376 pKa = 4.37GSGVRR1381 pKa = 11.84RR1382 pKa = 11.84AVAGNLALPLMMVGLVFNLL1401 pKa = 3.56
Molecular weight: 148.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2Z629|F2Z629_CANGA Ribosomal protein L37 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=CAGL0B01203g PE=3 SV=1
MM1 pKa = 7.82PAQKK5 pKa = 10.31SFKK8 pKa = 10.07IKK10 pKa = 10.38QKK12 pKa = 9.96LAKK15 pKa = 9.88AKK17 pKa = 9.79KK18 pKa = 8.92QNRR21 pKa = 11.84TLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.01WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.52MNII51 pKa = 3.1
MM1 pKa = 7.82PAQKK5 pKa = 10.31SFKK8 pKa = 10.07IKK10 pKa = 10.38QKK12 pKa = 9.96LAKK15 pKa = 9.88AKK17 pKa = 9.79KK18 pKa = 8.92QNRR21 pKa = 11.84TLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.01WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.52MNII51 pKa = 3.1
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2635782 |
34 |
4880 |
506.8 |
57.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.532 ± 0.031 | 1.161 ± 0.013 |
6.163 ± 0.028 | 6.76 ± 0.036 |
4.183 ± 0.025 | 5.247 ± 0.045 |
2.11 ± 0.013 | 6.523 ± 0.029 |
7.301 ± 0.035 | 9.315 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.211 ± 0.013 | 6.035 ± 0.028 |
4.25 ± 0.028 | 3.928 ± 0.027 |
4.435 ± 0.022 | 8.682 ± 0.058 |
5.852 ± 0.041 | 5.833 ± 0.024 |
0.991 ± 0.009 | 3.488 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
