
Torque teno mini virus 7
Taxonomy: Viruses; Anelloviridae; Betatorquevirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
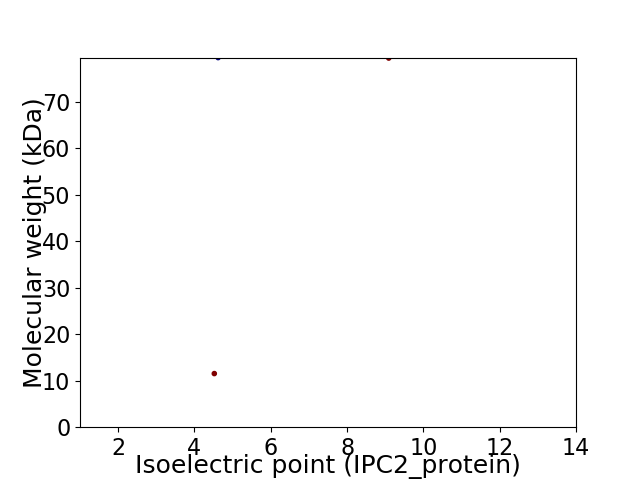
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9JG59|Q9JG59_9VIRU Uncharacterized protein OS=Torque teno mini virus 7 OX=687375 PE=4 SV=1
MM1 pKa = 7.71SRR3 pKa = 11.84QCPPKK8 pKa = 10.63YY9 pKa = 10.22NNKK12 pKa = 9.0QLQLQHH18 pKa = 6.35VNTLVSIHH26 pKa = 7.35DD27 pKa = 3.99LTCGCEE33 pKa = 4.2SPLEE37 pKa = 4.02CTIITIYY44 pKa = 10.61NQEE47 pKa = 4.0KK48 pKa = 10.12DD49 pKa = 3.54LKK51 pKa = 9.84FTEE54 pKa = 4.31EE55 pKa = 4.13EE56 pKa = 4.11KK57 pKa = 11.01KK58 pKa = 10.66KK59 pKa = 9.22LQKK62 pKa = 9.98WLTSTEE68 pKa = 3.97KK69 pKa = 10.35DD70 pKa = 3.29TTPEE74 pKa = 3.79EE75 pKa = 4.15EE76 pKa = 4.48GFGDD80 pKa = 4.14GDD82 pKa = 3.96LEE84 pKa = 4.46RR85 pKa = 11.84LFDD88 pKa = 4.49EE89 pKa = 5.95DD90 pKa = 5.26FGDD93 pKa = 5.0DD94 pKa = 3.75ADD96 pKa = 3.96TATRR100 pKa = 3.75
MM1 pKa = 7.71SRR3 pKa = 11.84QCPPKK8 pKa = 10.63YY9 pKa = 10.22NNKK12 pKa = 9.0QLQLQHH18 pKa = 6.35VNTLVSIHH26 pKa = 7.35DD27 pKa = 3.99LTCGCEE33 pKa = 4.2SPLEE37 pKa = 4.02CTIITIYY44 pKa = 10.61NQEE47 pKa = 4.0KK48 pKa = 10.12DD49 pKa = 3.54LKK51 pKa = 9.84FTEE54 pKa = 4.31EE55 pKa = 4.13EE56 pKa = 4.11KK57 pKa = 11.01KK58 pKa = 10.66KK59 pKa = 9.22LQKK62 pKa = 9.98WLTSTEE68 pKa = 3.97KK69 pKa = 10.35DD70 pKa = 3.29TTPEE74 pKa = 3.79EE75 pKa = 4.15EE76 pKa = 4.48GFGDD80 pKa = 4.14GDD82 pKa = 3.96LEE84 pKa = 4.46RR85 pKa = 11.84LFDD88 pKa = 4.49EE89 pKa = 5.95DD90 pKa = 5.26FGDD93 pKa = 5.0DD94 pKa = 3.75ADD96 pKa = 3.96TATRR100 pKa = 3.75
Molecular weight: 11.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9JG59|Q9JG59_9VIRU Uncharacterized protein OS=Torque teno mini virus 7 OX=687375 PE=4 SV=1
MM1 pKa = 7.72AYY3 pKa = 10.08FYY5 pKa = 10.84RR6 pKa = 11.84KK7 pKa = 9.08RR8 pKa = 11.84YY9 pKa = 7.24YY10 pKa = 9.25PRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FWRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84APFRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FWRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.07RR36 pKa = 11.84YY37 pKa = 9.15KK38 pKa = 10.59VKK40 pKa = 10.64RR41 pKa = 11.84KK42 pKa = 10.13LSTIKK47 pKa = 10.45LKK49 pKa = 10.1QWQPIYY55 pKa = 10.51LRR57 pKa = 11.84HH58 pKa = 5.83LKK60 pKa = 10.22IKK62 pKa = 10.68GILPLFITTDD72 pKa = 3.15KK73 pKa = 10.83RR74 pKa = 11.84ISNDD78 pKa = 2.62YY79 pKa = 10.18TMYY82 pKa = 10.38TNSLAQHH89 pKa = 5.68YY90 pKa = 9.7VPGGGGFTIYY100 pKa = 10.76RR101 pKa = 11.84FTLKK105 pKa = 10.61ALYY108 pKa = 10.19EE109 pKa = 4.46LFTKK113 pKa = 10.11IQNKK117 pKa = 5.56WTQSNEE123 pKa = 3.32DD124 pKa = 3.4WPFIRR129 pKa = 11.84YY130 pKa = 7.25TGCEE134 pKa = 3.68FKK136 pKa = 10.49FYY138 pKa = 10.62KK139 pKa = 10.0SEE141 pKa = 3.92KK142 pKa = 9.39ADD144 pKa = 3.72YY145 pKa = 9.82IINYY149 pKa = 7.78HH150 pKa = 6.47RR151 pKa = 11.84CPPMLATIDD160 pKa = 4.21TFHH163 pKa = 6.48STQPHH168 pKa = 5.59MMTLNKK174 pKa = 8.78KK175 pKa = 9.91HH176 pKa = 6.57KK177 pKa = 10.17IIRR180 pKa = 11.84CKK182 pKa = 10.32QNNPYY187 pKa = 9.35KK188 pKa = 10.43RR189 pKa = 11.84PYY191 pKa = 9.94KK192 pKa = 9.78RR193 pKa = 11.84VKK195 pKa = 9.97IKK197 pKa = 10.67PPALLQNNWYY207 pKa = 8.6FQKK210 pKa = 10.98DD211 pKa = 3.65FAAEE215 pKa = 3.86PLLLLMTSSMSLDD228 pKa = 3.0RR229 pKa = 11.84YY230 pKa = 9.61YY231 pKa = 11.35LNSKK235 pKa = 10.13SISTTMGFISLNDD248 pKa = 3.89NIFDD252 pKa = 3.9YY253 pKa = 11.31YY254 pKa = 11.05DD255 pKa = 3.54YY256 pKa = 11.19KK257 pKa = 11.15LPPTTGYY264 pKa = 9.8IPKK267 pKa = 8.91PKK269 pKa = 10.15KK270 pKa = 10.68YY271 pKa = 8.68MWSLYY276 pKa = 10.17QPLADD281 pKa = 3.4IHH283 pKa = 5.61KK284 pKa = 7.96TQIKK288 pKa = 9.37EE289 pKa = 3.92LIYY292 pKa = 10.41LGKK295 pKa = 10.16SKK297 pKa = 10.58EE298 pKa = 3.98MDD300 pKa = 2.8KK301 pKa = 11.4GKK303 pKa = 9.42PLKK306 pKa = 10.69NFLHH310 pKa = 6.98GDD312 pKa = 3.59PDD314 pKa = 4.12EE315 pKa = 4.22TWTNYY320 pKa = 9.44FSSTQNWGNIFDD332 pKa = 4.81PYY334 pKa = 11.21YY335 pKa = 9.51LTQEE339 pKa = 4.21HH340 pKa = 5.58VVLYY344 pKa = 10.09SEE346 pKa = 5.23KK347 pKa = 10.88SPNEE351 pKa = 3.81LKK353 pKa = 10.01TEE355 pKa = 4.0YY356 pKa = 10.61QKK358 pKa = 11.41DD359 pKa = 3.46KK360 pKa = 11.26QKK362 pKa = 10.72WKK364 pKa = 9.02TFSNEE369 pKa = 2.96IGEE372 pKa = 4.58YY373 pKa = 9.56IQEE376 pKa = 4.33TTHH379 pKa = 6.41TLLVNCRR386 pKa = 11.84YY387 pKa = 10.26NPYY390 pKa = 10.71ADD392 pKa = 3.34TGGNTVFIVPITQPEE407 pKa = 4.13QTPLHH412 pKa = 6.47IPQDD416 pKa = 3.88DD417 pKa = 4.06KK418 pKa = 11.9YY419 pKa = 9.81KK420 pKa = 10.2TEE422 pKa = 5.28PYY424 pKa = 9.88PLWLGTWGFLDD435 pKa = 3.96YY436 pKa = 10.55EE437 pKa = 4.47RR438 pKa = 11.84KK439 pKa = 10.23RR440 pKa = 11.84IGQTIDD446 pKa = 2.61TDD448 pKa = 3.79YY449 pKa = 11.38QIVIKK454 pKa = 10.27SPHH457 pKa = 5.82ITPSQYY463 pKa = 10.44FYY465 pKa = 11.09IPIDD469 pKa = 3.46EE470 pKa = 4.51TFTTGRR476 pKa = 11.84SPYY479 pKa = 9.73RR480 pKa = 11.84PPDD483 pKa = 3.8TQPTVYY489 pKa = 10.39DD490 pKa = 3.53QLHH493 pKa = 5.54WHH495 pKa = 6.83PKK497 pKa = 8.07ITMQMDD503 pKa = 4.91TINSIAACGPGTIKK517 pKa = 10.88LPPQVSAEE525 pKa = 4.03AHH527 pKa = 5.12CTYY530 pKa = 10.39TFHH533 pKa = 7.6FKK535 pKa = 10.65LGSCAPPMQNIDD547 pKa = 3.57NPEE550 pKa = 3.95KK551 pKa = 10.89QEE553 pKa = 3.96TFANNIFQTNSLQNPEE569 pKa = 4.62YY570 pKa = 9.59PLQYY574 pKa = 10.72YY575 pKa = 9.98LYY577 pKa = 10.54NFDD580 pKa = 4.37EE581 pKa = 4.29RR582 pKa = 11.84RR583 pKa = 11.84KK584 pKa = 9.34MLTKK588 pKa = 10.41RR589 pKa = 11.84AAKK592 pKa = 9.77RR593 pKa = 11.84IKK595 pKa = 10.36KK596 pKa = 10.16IIDD599 pKa = 3.52SEE601 pKa = 4.55KK602 pKa = 10.64SLSSTTGQSQLEE614 pKa = 4.14IPLQQEE620 pKa = 4.14AEE622 pKa = 3.99TDD624 pKa = 3.11ISSEE628 pKa = 4.02EE629 pKa = 4.01EE630 pKa = 4.08AEE632 pKa = 4.69KK633 pKa = 9.94EE634 pKa = 4.22TLLLLLRR641 pKa = 11.84QQRR644 pKa = 11.84LKK646 pKa = 9.2QAKK649 pKa = 8.33YY650 pKa = 10.03KK651 pKa = 10.37QRR653 pKa = 11.84LLQLIEE659 pKa = 3.87QLQTLEE665 pKa = 4.0
MM1 pKa = 7.72AYY3 pKa = 10.08FYY5 pKa = 10.84RR6 pKa = 11.84KK7 pKa = 9.08RR8 pKa = 11.84YY9 pKa = 7.24YY10 pKa = 9.25PRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84FWRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84PRR22 pKa = 11.84APFRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84FWRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84YY35 pKa = 9.07RR36 pKa = 11.84YY37 pKa = 9.15KK38 pKa = 10.59VKK40 pKa = 10.64RR41 pKa = 11.84KK42 pKa = 10.13LSTIKK47 pKa = 10.45LKK49 pKa = 10.1QWQPIYY55 pKa = 10.51LRR57 pKa = 11.84HH58 pKa = 5.83LKK60 pKa = 10.22IKK62 pKa = 10.68GILPLFITTDD72 pKa = 3.15KK73 pKa = 10.83RR74 pKa = 11.84ISNDD78 pKa = 2.62YY79 pKa = 10.18TMYY82 pKa = 10.38TNSLAQHH89 pKa = 5.68YY90 pKa = 9.7VPGGGGFTIYY100 pKa = 10.76RR101 pKa = 11.84FTLKK105 pKa = 10.61ALYY108 pKa = 10.19EE109 pKa = 4.46LFTKK113 pKa = 10.11IQNKK117 pKa = 5.56WTQSNEE123 pKa = 3.32DD124 pKa = 3.4WPFIRR129 pKa = 11.84YY130 pKa = 7.25TGCEE134 pKa = 3.68FKK136 pKa = 10.49FYY138 pKa = 10.62KK139 pKa = 10.0SEE141 pKa = 3.92KK142 pKa = 9.39ADD144 pKa = 3.72YY145 pKa = 9.82IINYY149 pKa = 7.78HH150 pKa = 6.47RR151 pKa = 11.84CPPMLATIDD160 pKa = 4.21TFHH163 pKa = 6.48STQPHH168 pKa = 5.59MMTLNKK174 pKa = 8.78KK175 pKa = 9.91HH176 pKa = 6.57KK177 pKa = 10.17IIRR180 pKa = 11.84CKK182 pKa = 10.32QNNPYY187 pKa = 9.35KK188 pKa = 10.43RR189 pKa = 11.84PYY191 pKa = 9.94KK192 pKa = 9.78RR193 pKa = 11.84VKK195 pKa = 9.97IKK197 pKa = 10.67PPALLQNNWYY207 pKa = 8.6FQKK210 pKa = 10.98DD211 pKa = 3.65FAAEE215 pKa = 3.86PLLLLMTSSMSLDD228 pKa = 3.0RR229 pKa = 11.84YY230 pKa = 9.61YY231 pKa = 11.35LNSKK235 pKa = 10.13SISTTMGFISLNDD248 pKa = 3.89NIFDD252 pKa = 3.9YY253 pKa = 11.31YY254 pKa = 11.05DD255 pKa = 3.54YY256 pKa = 11.19KK257 pKa = 11.15LPPTTGYY264 pKa = 9.8IPKK267 pKa = 8.91PKK269 pKa = 10.15KK270 pKa = 10.68YY271 pKa = 8.68MWSLYY276 pKa = 10.17QPLADD281 pKa = 3.4IHH283 pKa = 5.61KK284 pKa = 7.96TQIKK288 pKa = 9.37EE289 pKa = 3.92LIYY292 pKa = 10.41LGKK295 pKa = 10.16SKK297 pKa = 10.58EE298 pKa = 3.98MDD300 pKa = 2.8KK301 pKa = 11.4GKK303 pKa = 9.42PLKK306 pKa = 10.69NFLHH310 pKa = 6.98GDD312 pKa = 3.59PDD314 pKa = 4.12EE315 pKa = 4.22TWTNYY320 pKa = 9.44FSSTQNWGNIFDD332 pKa = 4.81PYY334 pKa = 11.21YY335 pKa = 9.51LTQEE339 pKa = 4.21HH340 pKa = 5.58VVLYY344 pKa = 10.09SEE346 pKa = 5.23KK347 pKa = 10.88SPNEE351 pKa = 3.81LKK353 pKa = 10.01TEE355 pKa = 4.0YY356 pKa = 10.61QKK358 pKa = 11.41DD359 pKa = 3.46KK360 pKa = 11.26QKK362 pKa = 10.72WKK364 pKa = 9.02TFSNEE369 pKa = 2.96IGEE372 pKa = 4.58YY373 pKa = 9.56IQEE376 pKa = 4.33TTHH379 pKa = 6.41TLLVNCRR386 pKa = 11.84YY387 pKa = 10.26NPYY390 pKa = 10.71ADD392 pKa = 3.34TGGNTVFIVPITQPEE407 pKa = 4.13QTPLHH412 pKa = 6.47IPQDD416 pKa = 3.88DD417 pKa = 4.06KK418 pKa = 11.9YY419 pKa = 9.81KK420 pKa = 10.2TEE422 pKa = 5.28PYY424 pKa = 9.88PLWLGTWGFLDD435 pKa = 3.96YY436 pKa = 10.55EE437 pKa = 4.47RR438 pKa = 11.84KK439 pKa = 10.23RR440 pKa = 11.84IGQTIDD446 pKa = 2.61TDD448 pKa = 3.79YY449 pKa = 11.38QIVIKK454 pKa = 10.27SPHH457 pKa = 5.82ITPSQYY463 pKa = 10.44FYY465 pKa = 11.09IPIDD469 pKa = 3.46EE470 pKa = 4.51TFTTGRR476 pKa = 11.84SPYY479 pKa = 9.73RR480 pKa = 11.84PPDD483 pKa = 3.8TQPTVYY489 pKa = 10.39DD490 pKa = 3.53QLHH493 pKa = 5.54WHH495 pKa = 6.83PKK497 pKa = 8.07ITMQMDD503 pKa = 4.91TINSIAACGPGTIKK517 pKa = 10.88LPPQVSAEE525 pKa = 4.03AHH527 pKa = 5.12CTYY530 pKa = 10.39TFHH533 pKa = 7.6FKK535 pKa = 10.65LGSCAPPMQNIDD547 pKa = 3.57NPEE550 pKa = 3.95KK551 pKa = 10.89QEE553 pKa = 3.96TFANNIFQTNSLQNPEE569 pKa = 4.62YY570 pKa = 9.59PLQYY574 pKa = 10.72YY575 pKa = 9.98LYY577 pKa = 10.54NFDD580 pKa = 4.37EE581 pKa = 4.29RR582 pKa = 11.84RR583 pKa = 11.84KK584 pKa = 9.34MLTKK588 pKa = 10.41RR589 pKa = 11.84AAKK592 pKa = 9.77RR593 pKa = 11.84IKK595 pKa = 10.36KK596 pKa = 10.16IIDD599 pKa = 3.52SEE601 pKa = 4.55KK602 pKa = 10.64SLSSTTGQSQLEE614 pKa = 4.14IPLQQEE620 pKa = 4.14AEE622 pKa = 3.99TDD624 pKa = 3.11ISSEE628 pKa = 4.02EE629 pKa = 4.01EE630 pKa = 4.08AEE632 pKa = 4.69KK633 pKa = 9.94EE634 pKa = 4.22TLLLLLRR641 pKa = 11.84QQRR644 pKa = 11.84LKK646 pKa = 9.2QAKK649 pKa = 8.33YY650 pKa = 10.03KK651 pKa = 10.37QRR653 pKa = 11.84LLQLIEE659 pKa = 3.87QLQTLEE665 pKa = 4.0
Molecular weight: 79.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
765 |
100 |
665 |
382.5 |
45.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.137 ± 0.497 | 1.438 ± 1.119 |
5.229 ± 2.084 | 6.144 ± 2.558 |
4.314 ± 0.137 | 3.66 ± 0.585 |
2.353 ± 0.154 | 6.667 ± 1.165 |
9.02 ± 0.009 | 8.889 ± 0.485 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.961 ± 0.42 | 4.444 ± 0.194 |
6.797 ± 1.222 | 6.275 ± 0.12 |
5.621 ± 1.145 | 4.967 ± 0.423 |
8.758 ± 0.979 | 1.699 ± 0.131 |
1.83 ± 0.363 | 6.797 ± 2.096 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
