
Leptosphaeria biglobosa mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 8.48
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4SUF2|A0A2R4SUF2_9VIRU RNA-dependent RNA polymerase OS=Leptosphaeria biglobosa mitovirus 1 OX=2163916 PE=4 SV=1
MM1 pKa = 7.41SLLFKK6 pKa = 10.66ILITMKK12 pKa = 10.39QNNYY16 pKa = 10.3LILEE20 pKa = 4.41RR21 pKa = 11.84LLKK24 pKa = 10.73SLFKK28 pKa = 10.01IDD30 pKa = 3.74CFAIVHH36 pKa = 5.81QYY38 pKa = 11.26LNYY41 pKa = 8.75IDD43 pKa = 5.25RR44 pKa = 11.84MRR46 pKa = 11.84EE47 pKa = 4.05KK48 pKa = 10.86NGLAYY53 pKa = 9.21TIKK56 pKa = 10.38HH57 pKa = 4.93MKK59 pKa = 8.74VVKK62 pKa = 10.05LHH64 pKa = 5.08ITRR67 pKa = 11.84FICNKK72 pKa = 8.95PLKK75 pKa = 10.46SNSNYY80 pKa = 9.93VSLDD84 pKa = 2.75KK85 pKa = 11.03DD86 pKa = 3.75YY87 pKa = 11.08FPSRR91 pKa = 11.84FLYY94 pKa = 10.46LKK96 pKa = 9.88RR97 pKa = 11.84FCKK100 pKa = 10.0TNPDD104 pKa = 3.4LVLTLLSYY112 pKa = 10.33TRR114 pKa = 11.84ALTPNKK120 pKa = 10.2RR121 pKa = 11.84EE122 pKa = 3.95TNARR126 pKa = 11.84NVDD129 pKa = 2.94ISSITNPYY137 pKa = 8.86KK138 pKa = 10.92GKK140 pKa = 10.48DD141 pKa = 3.3YY142 pKa = 10.11TIPIWFIKK150 pKa = 10.54DD151 pKa = 4.08FINHH155 pKa = 6.74HH156 pKa = 6.1GLSLSKK162 pKa = 10.11PVYY165 pKa = 10.45SNSDD169 pKa = 3.0HH170 pKa = 6.18YY171 pKa = 11.57LSIKK175 pKa = 10.22GSPNGKK181 pKa = 9.87SSISSLWSVASHH193 pKa = 6.7NEE195 pKa = 3.45NTLEE199 pKa = 4.19YY200 pKa = 9.92IRR202 pKa = 11.84TITGEE207 pKa = 3.92FFKK210 pKa = 10.77EE211 pKa = 3.35ISNYY215 pKa = 5.58YY216 pKa = 8.51TKK218 pKa = 10.44IVLDD222 pKa = 4.01YY223 pKa = 11.53SHH225 pKa = 7.4LIDD228 pKa = 4.03NNKK231 pKa = 9.95KK232 pKa = 9.56ILGKK236 pKa = 10.4LSVVHH241 pKa = 7.19DD242 pKa = 4.35PEE244 pKa = 4.86LKK246 pKa = 9.79EE247 pKa = 4.09RR248 pKa = 11.84VIAMVDD254 pKa = 3.27YY255 pKa = 7.64TTQFTLKK262 pKa = 10.0PIHH265 pKa = 5.96NQLLKK270 pKa = 10.89LLSKK274 pKa = 10.16FDD276 pKa = 3.93CDD278 pKa = 3.34RR279 pKa = 11.84TFTQDD284 pKa = 3.97PFHH287 pKa = 6.72NWQCNSEE294 pKa = 4.28NFHH297 pKa = 7.78SLDD300 pKa = 3.55LSAATDD306 pKa = 3.92RR307 pKa = 11.84FPIDD311 pKa = 3.68LQEE314 pKa = 4.45KK315 pKa = 8.53LLSYY319 pKa = 10.45IYY321 pKa = 10.03DD322 pKa = 3.92DD323 pKa = 4.77KK324 pKa = 11.82DD325 pKa = 3.49FAKK328 pKa = 10.01SWRR331 pKa = 11.84NLLVDD336 pKa = 3.83RR337 pKa = 11.84DD338 pKa = 4.51FIYY341 pKa = 9.8EE342 pKa = 4.07QKK344 pKa = 10.5SIRR347 pKa = 11.84YY348 pKa = 8.98SVGQPMGAYY357 pKa = 8.28TSWAAFTLTHH367 pKa = 6.32HH368 pKa = 7.16LVVQWAAKK376 pKa = 10.32LCGLDD381 pKa = 4.34HH382 pKa = 6.8FNQYY386 pKa = 10.4IILGDD391 pKa = 4.36DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.52YY404 pKa = 7.41ITIMTRR410 pKa = 11.84LGVDD414 pKa = 3.41ISQPKK419 pKa = 7.67THH421 pKa = 6.23VSNDD425 pKa = 3.06TYY427 pKa = 11.38EE428 pKa = 4.02FAKK431 pKa = 10.35RR432 pKa = 11.84WIQKK436 pKa = 9.22GRR438 pKa = 11.84EE439 pKa = 3.81ISGLSLKK446 pKa = 10.9GILNNFKK453 pKa = 10.29NKK455 pKa = 9.71HH456 pKa = 4.16VVYY459 pKa = 10.13MNIFNYY465 pKa = 9.12YY466 pKa = 9.61LRR468 pKa = 11.84RR469 pKa = 11.84PHH471 pKa = 8.32LDD473 pKa = 2.95IDD475 pKa = 4.12LLNLIGNLYY484 pKa = 10.41EE485 pKa = 4.76GIRR488 pKa = 11.84VSNRR492 pKa = 11.84IKK494 pKa = 10.83SKK496 pKa = 9.08NTIIKK501 pKa = 10.03LLYY504 pKa = 10.03DD505 pKa = 3.53FHH507 pKa = 8.45HH508 pKa = 7.19SIRR511 pKa = 11.84FSFNLLSYY519 pKa = 10.93EE520 pKa = 5.41DD521 pKa = 3.2IRR523 pKa = 11.84KK524 pKa = 9.33YY525 pKa = 11.07LNNKK529 pKa = 8.76IVIEE533 pKa = 4.25EE534 pKa = 4.08FNVWPEE540 pKa = 3.5SLIPYY545 pKa = 9.33KK546 pKa = 10.3IRR548 pKa = 11.84EE549 pKa = 4.01ILSYY553 pKa = 11.24GMVPQAEE560 pKa = 4.25SLKK563 pKa = 10.06TNISKK568 pKa = 10.26QMSILRR574 pKa = 11.84EE575 pKa = 4.21SKK577 pKa = 10.58DD578 pKa = 3.23IPIDD582 pKa = 4.32LLVDD586 pKa = 3.72WPLTYY591 pKa = 10.63GFKK594 pKa = 10.7NHH596 pKa = 6.61IEE598 pKa = 4.13SMTNLIEE605 pKa = 4.43GFVNKK610 pKa = 9.68TSQLLDD616 pKa = 4.04IISHH620 pKa = 6.38LRR622 pKa = 11.84LNNLDD627 pKa = 4.49GIMKK631 pKa = 10.22SRR633 pKa = 11.84LNNQDD638 pKa = 4.76LLLLDD643 pKa = 5.01KK644 pKa = 10.6LWKK647 pKa = 10.4DD648 pKa = 3.23SFSRR652 pKa = 11.84YY653 pKa = 9.06ILALKK658 pKa = 8.49TEE660 pKa = 4.33KK661 pKa = 10.95EE662 pKa = 4.24EE663 pKa = 4.11ILNSPEE669 pKa = 5.0DD670 pKa = 3.65KK671 pKa = 10.89PKK673 pKa = 11.21DD674 pKa = 3.68LVSHH678 pKa = 6.37IWSMAWASTPVVYY691 pKa = 8.69KK692 pKa = 10.26TSFVGLKK699 pKa = 10.34SFEE702 pKa = 4.42LYY704 pKa = 10.41LYY706 pKa = 10.74EE707 pKa = 5.91SFDD710 pKa = 4.45GLNTDD715 pKa = 3.74LDD717 pKa = 4.12YY718 pKa = 11.64LRR720 pKa = 11.84LEE722 pKa = 4.26TRR724 pKa = 11.84VDD726 pKa = 3.46EE727 pKa = 4.62SSTNIQDD734 pKa = 4.31RR735 pKa = 11.84IHH737 pKa = 7.03SFVYY741 pKa = 10.51DD742 pKa = 3.61SIKK745 pKa = 10.76NNKK748 pKa = 8.67HH749 pKa = 5.71INWDD753 pKa = 3.51EE754 pKa = 4.24FKK756 pKa = 11.19
MM1 pKa = 7.41SLLFKK6 pKa = 10.66ILITMKK12 pKa = 10.39QNNYY16 pKa = 10.3LILEE20 pKa = 4.41RR21 pKa = 11.84LLKK24 pKa = 10.73SLFKK28 pKa = 10.01IDD30 pKa = 3.74CFAIVHH36 pKa = 5.81QYY38 pKa = 11.26LNYY41 pKa = 8.75IDD43 pKa = 5.25RR44 pKa = 11.84MRR46 pKa = 11.84EE47 pKa = 4.05KK48 pKa = 10.86NGLAYY53 pKa = 9.21TIKK56 pKa = 10.38HH57 pKa = 4.93MKK59 pKa = 8.74VVKK62 pKa = 10.05LHH64 pKa = 5.08ITRR67 pKa = 11.84FICNKK72 pKa = 8.95PLKK75 pKa = 10.46SNSNYY80 pKa = 9.93VSLDD84 pKa = 2.75KK85 pKa = 11.03DD86 pKa = 3.75YY87 pKa = 11.08FPSRR91 pKa = 11.84FLYY94 pKa = 10.46LKK96 pKa = 9.88RR97 pKa = 11.84FCKK100 pKa = 10.0TNPDD104 pKa = 3.4LVLTLLSYY112 pKa = 10.33TRR114 pKa = 11.84ALTPNKK120 pKa = 10.2RR121 pKa = 11.84EE122 pKa = 3.95TNARR126 pKa = 11.84NVDD129 pKa = 2.94ISSITNPYY137 pKa = 8.86KK138 pKa = 10.92GKK140 pKa = 10.48DD141 pKa = 3.3YY142 pKa = 10.11TIPIWFIKK150 pKa = 10.54DD151 pKa = 4.08FINHH155 pKa = 6.74HH156 pKa = 6.1GLSLSKK162 pKa = 10.11PVYY165 pKa = 10.45SNSDD169 pKa = 3.0HH170 pKa = 6.18YY171 pKa = 11.57LSIKK175 pKa = 10.22GSPNGKK181 pKa = 9.87SSISSLWSVASHH193 pKa = 6.7NEE195 pKa = 3.45NTLEE199 pKa = 4.19YY200 pKa = 9.92IRR202 pKa = 11.84TITGEE207 pKa = 3.92FFKK210 pKa = 10.77EE211 pKa = 3.35ISNYY215 pKa = 5.58YY216 pKa = 8.51TKK218 pKa = 10.44IVLDD222 pKa = 4.01YY223 pKa = 11.53SHH225 pKa = 7.4LIDD228 pKa = 4.03NNKK231 pKa = 9.95KK232 pKa = 9.56ILGKK236 pKa = 10.4LSVVHH241 pKa = 7.19DD242 pKa = 4.35PEE244 pKa = 4.86LKK246 pKa = 9.79EE247 pKa = 4.09RR248 pKa = 11.84VIAMVDD254 pKa = 3.27YY255 pKa = 7.64TTQFTLKK262 pKa = 10.0PIHH265 pKa = 5.96NQLLKK270 pKa = 10.89LLSKK274 pKa = 10.16FDD276 pKa = 3.93CDD278 pKa = 3.34RR279 pKa = 11.84TFTQDD284 pKa = 3.97PFHH287 pKa = 6.72NWQCNSEE294 pKa = 4.28NFHH297 pKa = 7.78SLDD300 pKa = 3.55LSAATDD306 pKa = 3.92RR307 pKa = 11.84FPIDD311 pKa = 3.68LQEE314 pKa = 4.45KK315 pKa = 8.53LLSYY319 pKa = 10.45IYY321 pKa = 10.03DD322 pKa = 3.92DD323 pKa = 4.77KK324 pKa = 11.82DD325 pKa = 3.49FAKK328 pKa = 10.01SWRR331 pKa = 11.84NLLVDD336 pKa = 3.83RR337 pKa = 11.84DD338 pKa = 4.51FIYY341 pKa = 9.8EE342 pKa = 4.07QKK344 pKa = 10.5SIRR347 pKa = 11.84YY348 pKa = 8.98SVGQPMGAYY357 pKa = 8.28TSWAAFTLTHH367 pKa = 6.32HH368 pKa = 7.16LVVQWAAKK376 pKa = 10.32LCGLDD381 pKa = 4.34HH382 pKa = 6.8FNQYY386 pKa = 10.4IILGDD391 pKa = 4.36DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.52YY404 pKa = 7.41ITIMTRR410 pKa = 11.84LGVDD414 pKa = 3.41ISQPKK419 pKa = 7.67THH421 pKa = 6.23VSNDD425 pKa = 3.06TYY427 pKa = 11.38EE428 pKa = 4.02FAKK431 pKa = 10.35RR432 pKa = 11.84WIQKK436 pKa = 9.22GRR438 pKa = 11.84EE439 pKa = 3.81ISGLSLKK446 pKa = 10.9GILNNFKK453 pKa = 10.29NKK455 pKa = 9.71HH456 pKa = 4.16VVYY459 pKa = 10.13MNIFNYY465 pKa = 9.12YY466 pKa = 9.61LRR468 pKa = 11.84RR469 pKa = 11.84PHH471 pKa = 8.32LDD473 pKa = 2.95IDD475 pKa = 4.12LLNLIGNLYY484 pKa = 10.41EE485 pKa = 4.76GIRR488 pKa = 11.84VSNRR492 pKa = 11.84IKK494 pKa = 10.83SKK496 pKa = 9.08NTIIKK501 pKa = 10.03LLYY504 pKa = 10.03DD505 pKa = 3.53FHH507 pKa = 8.45HH508 pKa = 7.19SIRR511 pKa = 11.84FSFNLLSYY519 pKa = 10.93EE520 pKa = 5.41DD521 pKa = 3.2IRR523 pKa = 11.84KK524 pKa = 9.33YY525 pKa = 11.07LNNKK529 pKa = 8.76IVIEE533 pKa = 4.25EE534 pKa = 4.08FNVWPEE540 pKa = 3.5SLIPYY545 pKa = 9.33KK546 pKa = 10.3IRR548 pKa = 11.84EE549 pKa = 4.01ILSYY553 pKa = 11.24GMVPQAEE560 pKa = 4.25SLKK563 pKa = 10.06TNISKK568 pKa = 10.26QMSILRR574 pKa = 11.84EE575 pKa = 4.21SKK577 pKa = 10.58DD578 pKa = 3.23IPIDD582 pKa = 4.32LLVDD586 pKa = 3.72WPLTYY591 pKa = 10.63GFKK594 pKa = 10.7NHH596 pKa = 6.61IEE598 pKa = 4.13SMTNLIEE605 pKa = 4.43GFVNKK610 pKa = 9.68TSQLLDD616 pKa = 4.04IISHH620 pKa = 6.38LRR622 pKa = 11.84LNNLDD627 pKa = 4.49GIMKK631 pKa = 10.22SRR633 pKa = 11.84LNNQDD638 pKa = 4.76LLLLDD643 pKa = 5.01KK644 pKa = 10.6LWKK647 pKa = 10.4DD648 pKa = 3.23SFSRR652 pKa = 11.84YY653 pKa = 9.06ILALKK658 pKa = 8.49TEE660 pKa = 4.33KK661 pKa = 10.95EE662 pKa = 4.24EE663 pKa = 4.11ILNSPEE669 pKa = 5.0DD670 pKa = 3.65KK671 pKa = 10.89PKK673 pKa = 11.21DD674 pKa = 3.68LVSHH678 pKa = 6.37IWSMAWASTPVVYY691 pKa = 8.69KK692 pKa = 10.26TSFVGLKK699 pKa = 10.34SFEE702 pKa = 4.42LYY704 pKa = 10.41LYY706 pKa = 10.74EE707 pKa = 5.91SFDD710 pKa = 4.45GLNTDD715 pKa = 3.74LDD717 pKa = 4.12YY718 pKa = 11.64LRR720 pKa = 11.84LEE722 pKa = 4.26TRR724 pKa = 11.84VDD726 pKa = 3.46EE727 pKa = 4.62SSTNIQDD734 pKa = 4.31RR735 pKa = 11.84IHH737 pKa = 7.03SFVYY741 pKa = 10.51DD742 pKa = 3.61SIKK745 pKa = 10.76NNKK748 pKa = 8.67HH749 pKa = 5.71INWDD753 pKa = 3.51EE754 pKa = 4.24FKK756 pKa = 11.19
Molecular weight: 88.78 kDa
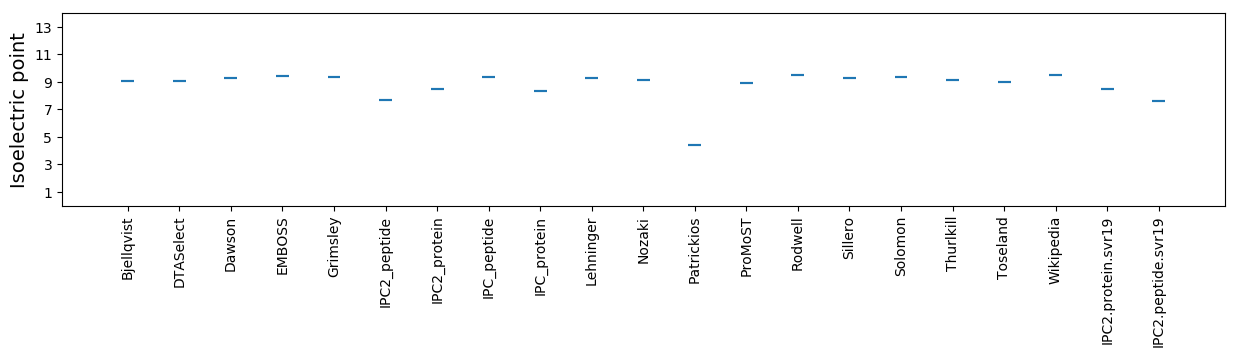
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4SUF2|A0A2R4SUF2_9VIRU RNA-dependent RNA polymerase OS=Leptosphaeria biglobosa mitovirus 1 OX=2163916 PE=4 SV=1
MM1 pKa = 7.41SLLFKK6 pKa = 10.66ILITMKK12 pKa = 10.39QNNYY16 pKa = 10.3LILEE20 pKa = 4.41RR21 pKa = 11.84LLKK24 pKa = 10.73SLFKK28 pKa = 10.01IDD30 pKa = 3.74CFAIVHH36 pKa = 5.81QYY38 pKa = 11.26LNYY41 pKa = 8.75IDD43 pKa = 5.25RR44 pKa = 11.84MRR46 pKa = 11.84EE47 pKa = 4.05KK48 pKa = 10.86NGLAYY53 pKa = 9.21TIKK56 pKa = 10.38HH57 pKa = 4.93MKK59 pKa = 8.74VVKK62 pKa = 10.05LHH64 pKa = 5.08ITRR67 pKa = 11.84FICNKK72 pKa = 8.95PLKK75 pKa = 10.46SNSNYY80 pKa = 9.93VSLDD84 pKa = 2.75KK85 pKa = 11.03DD86 pKa = 3.75YY87 pKa = 11.08FPSRR91 pKa = 11.84FLYY94 pKa = 10.46LKK96 pKa = 9.88RR97 pKa = 11.84FCKK100 pKa = 10.0TNPDD104 pKa = 3.4LVLTLLSYY112 pKa = 10.33TRR114 pKa = 11.84ALTPNKK120 pKa = 10.2RR121 pKa = 11.84EE122 pKa = 3.95TNARR126 pKa = 11.84NVDD129 pKa = 2.94ISSITNPYY137 pKa = 8.86KK138 pKa = 10.92GKK140 pKa = 10.48DD141 pKa = 3.3YY142 pKa = 10.11TIPIWFIKK150 pKa = 10.54DD151 pKa = 4.08FINHH155 pKa = 6.74HH156 pKa = 6.1GLSLSKK162 pKa = 10.11PVYY165 pKa = 10.45SNSDD169 pKa = 3.0HH170 pKa = 6.18YY171 pKa = 11.57LSIKK175 pKa = 10.22GSPNGKK181 pKa = 9.87SSISSLWSVASHH193 pKa = 6.7NEE195 pKa = 3.45NTLEE199 pKa = 4.19YY200 pKa = 9.92IRR202 pKa = 11.84TITGEE207 pKa = 3.92FFKK210 pKa = 10.77EE211 pKa = 3.35ISNYY215 pKa = 5.58YY216 pKa = 8.51TKK218 pKa = 10.44IVLDD222 pKa = 4.01YY223 pKa = 11.53SHH225 pKa = 7.4LIDD228 pKa = 4.03NNKK231 pKa = 9.95KK232 pKa = 9.56ILGKK236 pKa = 10.4LSVVHH241 pKa = 7.19DD242 pKa = 4.35PEE244 pKa = 4.86LKK246 pKa = 9.79EE247 pKa = 4.09RR248 pKa = 11.84VIAMVDD254 pKa = 3.27YY255 pKa = 7.64TTQFTLKK262 pKa = 10.0PIHH265 pKa = 5.96NQLLKK270 pKa = 10.89LLSKK274 pKa = 10.16FDD276 pKa = 3.93CDD278 pKa = 3.34RR279 pKa = 11.84TFTQDD284 pKa = 3.97PFHH287 pKa = 6.72NWQCNSEE294 pKa = 4.28NFHH297 pKa = 7.78SLDD300 pKa = 3.55LSAATDD306 pKa = 3.92RR307 pKa = 11.84FPIDD311 pKa = 3.68LQEE314 pKa = 4.45KK315 pKa = 8.53LLSYY319 pKa = 10.45IYY321 pKa = 10.03DD322 pKa = 3.92DD323 pKa = 4.77KK324 pKa = 11.82DD325 pKa = 3.49FAKK328 pKa = 10.01SWRR331 pKa = 11.84NLLVDD336 pKa = 3.83RR337 pKa = 11.84DD338 pKa = 4.51FIYY341 pKa = 9.8EE342 pKa = 4.07QKK344 pKa = 10.5SIRR347 pKa = 11.84YY348 pKa = 8.98SVGQPMGAYY357 pKa = 8.28TSWAAFTLTHH367 pKa = 6.32HH368 pKa = 7.16LVVQWAAKK376 pKa = 10.32LCGLDD381 pKa = 4.34HH382 pKa = 6.8FNQYY386 pKa = 10.4IILGDD391 pKa = 4.36DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.52YY404 pKa = 7.41ITIMTRR410 pKa = 11.84LGVDD414 pKa = 3.41ISQPKK419 pKa = 7.67THH421 pKa = 6.23VSNDD425 pKa = 3.06TYY427 pKa = 11.38EE428 pKa = 4.02FAKK431 pKa = 10.35RR432 pKa = 11.84WIQKK436 pKa = 9.22GRR438 pKa = 11.84EE439 pKa = 3.81ISGLSLKK446 pKa = 10.9GILNNFKK453 pKa = 10.29NKK455 pKa = 9.71HH456 pKa = 4.16VVYY459 pKa = 10.13MNIFNYY465 pKa = 9.12YY466 pKa = 9.61LRR468 pKa = 11.84RR469 pKa = 11.84PHH471 pKa = 8.32LDD473 pKa = 2.95IDD475 pKa = 4.12LLNLIGNLYY484 pKa = 10.41EE485 pKa = 4.76GIRR488 pKa = 11.84VSNRR492 pKa = 11.84IKK494 pKa = 10.83SKK496 pKa = 9.08NTIIKK501 pKa = 10.03LLYY504 pKa = 10.03DD505 pKa = 3.53FHH507 pKa = 8.45HH508 pKa = 7.19SIRR511 pKa = 11.84FSFNLLSYY519 pKa = 10.93EE520 pKa = 5.41DD521 pKa = 3.2IRR523 pKa = 11.84KK524 pKa = 9.33YY525 pKa = 11.07LNNKK529 pKa = 8.76IVIEE533 pKa = 4.25EE534 pKa = 4.08FNVWPEE540 pKa = 3.5SLIPYY545 pKa = 9.33KK546 pKa = 10.3IRR548 pKa = 11.84EE549 pKa = 4.01ILSYY553 pKa = 11.24GMVPQAEE560 pKa = 4.25SLKK563 pKa = 10.06TNISKK568 pKa = 10.26QMSILRR574 pKa = 11.84EE575 pKa = 4.21SKK577 pKa = 10.58DD578 pKa = 3.23IPIDD582 pKa = 4.32LLVDD586 pKa = 3.72WPLTYY591 pKa = 10.63GFKK594 pKa = 10.7NHH596 pKa = 6.61IEE598 pKa = 4.13SMTNLIEE605 pKa = 4.43GFVNKK610 pKa = 9.68TSQLLDD616 pKa = 4.04IISHH620 pKa = 6.38LRR622 pKa = 11.84LNNLDD627 pKa = 4.49GIMKK631 pKa = 10.22SRR633 pKa = 11.84LNNQDD638 pKa = 4.76LLLLDD643 pKa = 5.01KK644 pKa = 10.6LWKK647 pKa = 10.4DD648 pKa = 3.23SFSRR652 pKa = 11.84YY653 pKa = 9.06ILALKK658 pKa = 8.49TEE660 pKa = 4.33KK661 pKa = 10.95EE662 pKa = 4.24EE663 pKa = 4.11ILNSPEE669 pKa = 5.0DD670 pKa = 3.65KK671 pKa = 10.89PKK673 pKa = 11.21DD674 pKa = 3.68LVSHH678 pKa = 6.37IWSMAWASTPVVYY691 pKa = 8.69KK692 pKa = 10.26TSFVGLKK699 pKa = 10.34SFEE702 pKa = 4.42LYY704 pKa = 10.41LYY706 pKa = 10.74EE707 pKa = 5.91SFDD710 pKa = 4.45GLNTDD715 pKa = 3.74LDD717 pKa = 4.12YY718 pKa = 11.64LRR720 pKa = 11.84LEE722 pKa = 4.26TRR724 pKa = 11.84VDD726 pKa = 3.46EE727 pKa = 4.62SSTNIQDD734 pKa = 4.31RR735 pKa = 11.84IHH737 pKa = 7.03SFVYY741 pKa = 10.51DD742 pKa = 3.61SIKK745 pKa = 10.76NNKK748 pKa = 8.67HH749 pKa = 5.71INWDD753 pKa = 3.51EE754 pKa = 4.24FKK756 pKa = 11.19
MM1 pKa = 7.41SLLFKK6 pKa = 10.66ILITMKK12 pKa = 10.39QNNYY16 pKa = 10.3LILEE20 pKa = 4.41RR21 pKa = 11.84LLKK24 pKa = 10.73SLFKK28 pKa = 10.01IDD30 pKa = 3.74CFAIVHH36 pKa = 5.81QYY38 pKa = 11.26LNYY41 pKa = 8.75IDD43 pKa = 5.25RR44 pKa = 11.84MRR46 pKa = 11.84EE47 pKa = 4.05KK48 pKa = 10.86NGLAYY53 pKa = 9.21TIKK56 pKa = 10.38HH57 pKa = 4.93MKK59 pKa = 8.74VVKK62 pKa = 10.05LHH64 pKa = 5.08ITRR67 pKa = 11.84FICNKK72 pKa = 8.95PLKK75 pKa = 10.46SNSNYY80 pKa = 9.93VSLDD84 pKa = 2.75KK85 pKa = 11.03DD86 pKa = 3.75YY87 pKa = 11.08FPSRR91 pKa = 11.84FLYY94 pKa = 10.46LKK96 pKa = 9.88RR97 pKa = 11.84FCKK100 pKa = 10.0TNPDD104 pKa = 3.4LVLTLLSYY112 pKa = 10.33TRR114 pKa = 11.84ALTPNKK120 pKa = 10.2RR121 pKa = 11.84EE122 pKa = 3.95TNARR126 pKa = 11.84NVDD129 pKa = 2.94ISSITNPYY137 pKa = 8.86KK138 pKa = 10.92GKK140 pKa = 10.48DD141 pKa = 3.3YY142 pKa = 10.11TIPIWFIKK150 pKa = 10.54DD151 pKa = 4.08FINHH155 pKa = 6.74HH156 pKa = 6.1GLSLSKK162 pKa = 10.11PVYY165 pKa = 10.45SNSDD169 pKa = 3.0HH170 pKa = 6.18YY171 pKa = 11.57LSIKK175 pKa = 10.22GSPNGKK181 pKa = 9.87SSISSLWSVASHH193 pKa = 6.7NEE195 pKa = 3.45NTLEE199 pKa = 4.19YY200 pKa = 9.92IRR202 pKa = 11.84TITGEE207 pKa = 3.92FFKK210 pKa = 10.77EE211 pKa = 3.35ISNYY215 pKa = 5.58YY216 pKa = 8.51TKK218 pKa = 10.44IVLDD222 pKa = 4.01YY223 pKa = 11.53SHH225 pKa = 7.4LIDD228 pKa = 4.03NNKK231 pKa = 9.95KK232 pKa = 9.56ILGKK236 pKa = 10.4LSVVHH241 pKa = 7.19DD242 pKa = 4.35PEE244 pKa = 4.86LKK246 pKa = 9.79EE247 pKa = 4.09RR248 pKa = 11.84VIAMVDD254 pKa = 3.27YY255 pKa = 7.64TTQFTLKK262 pKa = 10.0PIHH265 pKa = 5.96NQLLKK270 pKa = 10.89LLSKK274 pKa = 10.16FDD276 pKa = 3.93CDD278 pKa = 3.34RR279 pKa = 11.84TFTQDD284 pKa = 3.97PFHH287 pKa = 6.72NWQCNSEE294 pKa = 4.28NFHH297 pKa = 7.78SLDD300 pKa = 3.55LSAATDD306 pKa = 3.92RR307 pKa = 11.84FPIDD311 pKa = 3.68LQEE314 pKa = 4.45KK315 pKa = 8.53LLSYY319 pKa = 10.45IYY321 pKa = 10.03DD322 pKa = 3.92DD323 pKa = 4.77KK324 pKa = 11.82DD325 pKa = 3.49FAKK328 pKa = 10.01SWRR331 pKa = 11.84NLLVDD336 pKa = 3.83RR337 pKa = 11.84DD338 pKa = 4.51FIYY341 pKa = 9.8EE342 pKa = 4.07QKK344 pKa = 10.5SIRR347 pKa = 11.84YY348 pKa = 8.98SVGQPMGAYY357 pKa = 8.28TSWAAFTLTHH367 pKa = 6.32HH368 pKa = 7.16LVVQWAAKK376 pKa = 10.32LCGLDD381 pKa = 4.34HH382 pKa = 6.8FNQYY386 pKa = 10.4IILGDD391 pKa = 4.36DD392 pKa = 3.31IVIKK396 pKa = 10.53NNKK399 pKa = 7.97VANKK403 pKa = 9.52YY404 pKa = 7.41ITIMTRR410 pKa = 11.84LGVDD414 pKa = 3.41ISQPKK419 pKa = 7.67THH421 pKa = 6.23VSNDD425 pKa = 3.06TYY427 pKa = 11.38EE428 pKa = 4.02FAKK431 pKa = 10.35RR432 pKa = 11.84WIQKK436 pKa = 9.22GRR438 pKa = 11.84EE439 pKa = 3.81ISGLSLKK446 pKa = 10.9GILNNFKK453 pKa = 10.29NKK455 pKa = 9.71HH456 pKa = 4.16VVYY459 pKa = 10.13MNIFNYY465 pKa = 9.12YY466 pKa = 9.61LRR468 pKa = 11.84RR469 pKa = 11.84PHH471 pKa = 8.32LDD473 pKa = 2.95IDD475 pKa = 4.12LLNLIGNLYY484 pKa = 10.41EE485 pKa = 4.76GIRR488 pKa = 11.84VSNRR492 pKa = 11.84IKK494 pKa = 10.83SKK496 pKa = 9.08NTIIKK501 pKa = 10.03LLYY504 pKa = 10.03DD505 pKa = 3.53FHH507 pKa = 8.45HH508 pKa = 7.19SIRR511 pKa = 11.84FSFNLLSYY519 pKa = 10.93EE520 pKa = 5.41DD521 pKa = 3.2IRR523 pKa = 11.84KK524 pKa = 9.33YY525 pKa = 11.07LNNKK529 pKa = 8.76IVIEE533 pKa = 4.25EE534 pKa = 4.08FNVWPEE540 pKa = 3.5SLIPYY545 pKa = 9.33KK546 pKa = 10.3IRR548 pKa = 11.84EE549 pKa = 4.01ILSYY553 pKa = 11.24GMVPQAEE560 pKa = 4.25SLKK563 pKa = 10.06TNISKK568 pKa = 10.26QMSILRR574 pKa = 11.84EE575 pKa = 4.21SKK577 pKa = 10.58DD578 pKa = 3.23IPIDD582 pKa = 4.32LLVDD586 pKa = 3.72WPLTYY591 pKa = 10.63GFKK594 pKa = 10.7NHH596 pKa = 6.61IEE598 pKa = 4.13SMTNLIEE605 pKa = 4.43GFVNKK610 pKa = 9.68TSQLLDD616 pKa = 4.04IISHH620 pKa = 6.38LRR622 pKa = 11.84LNNLDD627 pKa = 4.49GIMKK631 pKa = 10.22SRR633 pKa = 11.84LNNQDD638 pKa = 4.76LLLLDD643 pKa = 5.01KK644 pKa = 10.6LWKK647 pKa = 10.4DD648 pKa = 3.23SFSRR652 pKa = 11.84YY653 pKa = 9.06ILALKK658 pKa = 8.49TEE660 pKa = 4.33KK661 pKa = 10.95EE662 pKa = 4.24EE663 pKa = 4.11ILNSPEE669 pKa = 5.0DD670 pKa = 3.65KK671 pKa = 10.89PKK673 pKa = 11.21DD674 pKa = 3.68LVSHH678 pKa = 6.37IWSMAWASTPVVYY691 pKa = 8.69KK692 pKa = 10.26TSFVGLKK699 pKa = 10.34SFEE702 pKa = 4.42LYY704 pKa = 10.41LYY706 pKa = 10.74EE707 pKa = 5.91SFDD710 pKa = 4.45GLNTDD715 pKa = 3.74LDD717 pKa = 4.12YY718 pKa = 11.64LRR720 pKa = 11.84LEE722 pKa = 4.26TRR724 pKa = 11.84VDD726 pKa = 3.46EE727 pKa = 4.62SSTNIQDD734 pKa = 4.31RR735 pKa = 11.84IHH737 pKa = 7.03SFVYY741 pKa = 10.51DD742 pKa = 3.61SIKK745 pKa = 10.76NNKK748 pKa = 8.67HH749 pKa = 5.71INWDD753 pKa = 3.51EE754 pKa = 4.24FKK756 pKa = 11.19
Molecular weight: 88.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
756 |
756 |
756 |
756.0 |
88.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.646 ± 0.0 | 0.794 ± 0.0 |
6.614 ± 0.0 | 4.365 ± 0.0 |
4.762 ± 0.0 | 3.042 ± 0.0 |
3.307 ± 0.0 | 9.127 ± 0.0 |
8.73 ± 0.0 | 12.169 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.72 ± 0.0 | 7.54 ± 0.0 |
3.042 ± 0.0 | 2.381 ± 0.0 |
4.365 ± 0.0 | 8.598 ± 0.0 |
5.026 ± 0.0 | 4.63 ± 0.0 |
1.72 ± 0.0 | 5.423 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
