
Wuhan house centipede virus 6
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
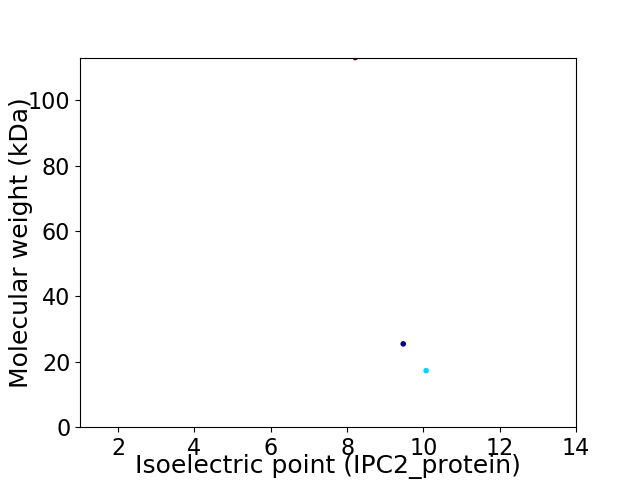
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGS6|A0A1L3KGS6_9VIRU Uncharacterized protein OS=Wuhan house centipede virus 6 OX=1923710 PE=4 SV=1
MM1 pKa = 7.29SQMLFNTCDD10 pKa = 3.45VVKK13 pKa = 10.08STFLMMRR20 pKa = 11.84TYY22 pKa = 10.53PKK24 pKa = 9.34TVLAITVTSYY34 pKa = 11.97GLVKK38 pKa = 10.56LIKK41 pKa = 10.16HH42 pKa = 5.59HH43 pKa = 6.88AKK45 pKa = 10.43LYY47 pKa = 8.12NQQNPFEE54 pKa = 4.68YY55 pKa = 10.14IPSEE59 pKa = 3.99EE60 pKa = 4.43TNWLLVRR67 pKa = 11.84QVRR70 pKa = 11.84NVLKK74 pKa = 10.2HH75 pKa = 5.04QYY77 pKa = 10.7AAVKK81 pKa = 8.49YY82 pKa = 10.61QMMTVGGLYY91 pKa = 7.79QTNVKK96 pKa = 9.81QLSEE100 pKa = 4.58EE101 pKa = 3.93IEE103 pKa = 3.89KK104 pKa = 10.16TIIRR108 pKa = 11.84KK109 pKa = 9.13PDD111 pKa = 3.41LRR113 pKa = 11.84PTLVPNGDD121 pKa = 4.36SIDD124 pKa = 4.21IIPVNSMHH132 pKa = 6.25SHH134 pKa = 6.55PASAQFRR141 pKa = 11.84SSANSYY147 pKa = 11.33LNDD150 pKa = 3.53CVVRR154 pKa = 11.84AGYY157 pKa = 9.99RR158 pKa = 11.84PYY160 pKa = 10.68NISKK164 pKa = 9.92SRR166 pKa = 11.84RR167 pKa = 11.84DD168 pKa = 3.3IEE170 pKa = 4.35DD171 pKa = 3.32GCRR174 pKa = 11.84YY175 pKa = 9.88FYY177 pKa = 10.64HH178 pKa = 7.18SKK180 pKa = 10.69DD181 pKa = 3.61LSIKK185 pKa = 9.95YY186 pKa = 10.62SNDD189 pKa = 3.28KK190 pKa = 11.0IEE192 pKa = 4.55DD193 pKa = 3.33QSVFIMTDD201 pKa = 2.15VDD203 pKa = 4.4YY204 pKa = 11.26YY205 pKa = 11.63VDD207 pKa = 3.46MPRR210 pKa = 11.84WLKK213 pKa = 10.38LFRR216 pKa = 11.84PIVMYY221 pKa = 9.65TLSPTKK227 pKa = 10.45LSCSGPNDD235 pKa = 3.3KK236 pKa = 11.17AEE238 pKa = 3.73YY239 pKa = 10.16RR240 pKa = 11.84FNIDD244 pKa = 3.05GNVVHH249 pKa = 5.97YY250 pKa = 9.81HH251 pKa = 4.66VSGGGEE257 pKa = 3.98YY258 pKa = 8.71THH260 pKa = 6.48QLWNYY265 pKa = 8.41TGDD268 pKa = 3.41TVTCVDD274 pKa = 4.64DD275 pKa = 5.6DD276 pKa = 4.96GNLLTFDD283 pKa = 3.4IEE285 pKa = 4.17QRR287 pKa = 11.84KK288 pKa = 9.09VKK290 pKa = 10.46GDD292 pKa = 3.48EE293 pKa = 3.68QHH295 pKa = 6.71RR296 pKa = 11.84LIWLLPRR303 pKa = 11.84SKK305 pKa = 9.45VTNPLWHH312 pKa = 6.53YY313 pKa = 11.1LRR315 pKa = 11.84LDD317 pKa = 3.41WEE319 pKa = 4.45NNLLKK324 pKa = 10.69RR325 pKa = 11.84KK326 pKa = 10.35DD327 pKa = 3.44MTLKK331 pKa = 10.5GFNYY335 pKa = 10.05LLEE338 pKa = 5.11PIEE341 pKa = 5.06DD342 pKa = 3.77NLSIGLIGTKK352 pKa = 10.29YY353 pKa = 10.44SVEE356 pKa = 4.14IPGKK360 pKa = 9.29LYY362 pKa = 9.55EE363 pKa = 4.73AIRR366 pKa = 11.84IRR368 pKa = 11.84LAEE371 pKa = 4.08KK372 pKa = 9.39EE373 pKa = 4.13ATVFVSDD380 pKa = 3.63VEE382 pKa = 4.6RR383 pKa = 11.84MLKK386 pKa = 9.04EE387 pKa = 4.35AKK389 pKa = 9.19HH390 pKa = 4.97QSYY393 pKa = 7.97VTDD396 pKa = 3.7APILFKK402 pKa = 10.99CFAVDD407 pKa = 3.89VVFDD411 pKa = 4.15KK412 pKa = 11.55NVVKK416 pKa = 10.09TSNFAVTYY424 pKa = 9.0QAIPRR429 pKa = 11.84HH430 pKa = 5.69GALSTEE436 pKa = 4.67DD437 pKa = 3.87GNNPGQTTTSPLTSNPALFASKK459 pKa = 10.56GYY461 pKa = 9.96NADD464 pKa = 3.52KK465 pKa = 11.05ACIEE469 pKa = 4.0GRR471 pKa = 11.84IHH473 pKa = 7.31KK474 pKa = 9.31VANNKK479 pKa = 8.84KK480 pKa = 8.52WSKK483 pKa = 10.36KK484 pKa = 6.7YY485 pKa = 10.1HH486 pKa = 6.39DD487 pKa = 4.1YY488 pKa = 11.25ARR490 pKa = 11.84EE491 pKa = 4.21FIDD494 pKa = 3.57RR495 pKa = 11.84LVPKK499 pKa = 9.76RR500 pKa = 11.84YY501 pKa = 9.68RR502 pKa = 11.84GTGTPLSIGEE512 pKa = 3.83VDD514 pKa = 3.64LRR516 pKa = 11.84QEE518 pKa = 3.99KK519 pKa = 9.2VAQRR523 pKa = 11.84GRR525 pKa = 11.84FKK527 pKa = 10.7QVAPMMSLDD536 pKa = 3.6AGNAIKK542 pKa = 10.75AFIKK546 pKa = 10.04TEE548 pKa = 4.23TYY550 pKa = 11.13ASAKK554 pKa = 8.85PPRR557 pKa = 11.84NISTMSPEE565 pKa = 3.97LTIQMSAYY573 pKa = 8.67TLVFANILKK582 pKa = 8.5THH584 pKa = 6.9DD585 pKa = 3.56WYY587 pKa = 11.54CPGKK591 pKa = 10.22KK592 pKa = 9.59PRR594 pKa = 11.84DD595 pKa = 3.49IIKK598 pKa = 10.53RR599 pKa = 11.84LAHH602 pKa = 5.07VMKK605 pKa = 10.73AEE607 pKa = 3.94EE608 pKa = 4.42TEE610 pKa = 4.08DD611 pKa = 3.96VEE613 pKa = 4.9EE614 pKa = 5.2GDD616 pKa = 4.15YY617 pKa = 11.08TCLDD621 pKa = 3.59GTQSEE626 pKa = 5.13EE627 pKa = 4.01YY628 pKa = 10.56ARR630 pKa = 11.84NLLLPAYY637 pKa = 7.78MQYY640 pKa = 10.98LAPEE644 pKa = 4.34HH645 pKa = 6.0RR646 pKa = 11.84TMFKK650 pKa = 10.78DD651 pKa = 4.22LYY653 pKa = 10.04KK654 pKa = 10.46KK655 pKa = 10.45VYY657 pKa = 8.61KK658 pKa = 10.29QHH660 pKa = 6.28ATTSTGVSYY669 pKa = 11.1DD670 pKa = 2.81PGYY673 pKa = 8.72TVRR676 pKa = 11.84SGSPITTQAGTLANAFNVYY695 pKa = 10.06SALRR699 pKa = 11.84NMGYY703 pKa = 10.15VPDD706 pKa = 4.46EE707 pKa = 4.36AFDD710 pKa = 3.84MIGAIFGDD718 pKa = 4.47DD719 pKa = 3.57SCNPNYY725 pKa = 10.15KK726 pKa = 10.39GQFHH730 pKa = 7.0LFIEE734 pKa = 4.74QVAKK738 pKa = 10.41DD739 pKa = 3.55LGMIYY744 pKa = 10.34KK745 pKa = 10.57SNLRR749 pKa = 11.84PRR751 pKa = 11.84GEE753 pKa = 3.9PLLFLGRR760 pKa = 11.84YY761 pKa = 8.42FVDD764 pKa = 4.86PPTTDD769 pKa = 4.07DD770 pKa = 4.5SYY772 pKa = 12.1ADD774 pKa = 3.72PLRR777 pKa = 11.84TIGKK781 pKa = 8.69LHH783 pKa = 6.91ASTNKK788 pKa = 10.22SVTKK792 pKa = 10.09EE793 pKa = 3.26QAAANKK799 pKa = 9.98ALGYY803 pKa = 10.55LSTDD807 pKa = 2.8RR808 pKa = 11.84LTPIIGTWAARR819 pKa = 11.84VIQLTKK825 pKa = 9.98IRR827 pKa = 11.84TMKK830 pKa = 10.64GGTGEE835 pKa = 4.06EE836 pKa = 4.31QYY838 pKa = 10.79KK839 pKa = 10.71CSNAWPQKK847 pKa = 9.55DD848 pKa = 3.27EE849 pKa = 4.05TRR851 pKa = 11.84IRR853 pKa = 11.84EE854 pKa = 4.26CMATTLGITTAEE866 pKa = 4.74LIEE869 pKa = 4.53LDD871 pKa = 3.52KK872 pKa = 10.96KK873 pKa = 10.5IRR875 pKa = 11.84SVDD878 pKa = 3.65GLDD881 pKa = 3.45HH882 pKa = 6.71FPVIFDD888 pKa = 3.39TAYY891 pKa = 9.9KK892 pKa = 7.8HH893 pKa = 5.02TQVAVVDD900 pKa = 4.11GDD902 pKa = 4.16LVCTDD907 pKa = 3.42LHH909 pKa = 7.72QDD911 pKa = 3.21IEE913 pKa = 4.78PIHH916 pKa = 7.06DD917 pKa = 3.59EE918 pKa = 4.42SEE920 pKa = 4.3PKK922 pKa = 10.43ASCSGIQSPSSSHH935 pKa = 6.3RR936 pKa = 11.84SCSAKK941 pKa = 9.48ATEE944 pKa = 4.16RR945 pKa = 11.84PKK947 pKa = 10.76PGFTEE952 pKa = 4.84RR953 pKa = 11.84SFKK956 pKa = 10.3TSGRR960 pKa = 11.84RR961 pKa = 11.84ATGKK965 pKa = 9.54GHH967 pKa = 7.1PNRR970 pKa = 11.84IRR972 pKa = 11.84PNTRR976 pKa = 11.84KK977 pKa = 10.04SNRR980 pKa = 11.84VDD982 pKa = 3.26PRR984 pKa = 11.84DD985 pKa = 3.75DD986 pKa = 3.44VPRR989 pKa = 11.84TDD991 pKa = 3.32GRR993 pKa = 11.84RR994 pKa = 11.84INN996 pKa = 3.54
MM1 pKa = 7.29SQMLFNTCDD10 pKa = 3.45VVKK13 pKa = 10.08STFLMMRR20 pKa = 11.84TYY22 pKa = 10.53PKK24 pKa = 9.34TVLAITVTSYY34 pKa = 11.97GLVKK38 pKa = 10.56LIKK41 pKa = 10.16HH42 pKa = 5.59HH43 pKa = 6.88AKK45 pKa = 10.43LYY47 pKa = 8.12NQQNPFEE54 pKa = 4.68YY55 pKa = 10.14IPSEE59 pKa = 3.99EE60 pKa = 4.43TNWLLVRR67 pKa = 11.84QVRR70 pKa = 11.84NVLKK74 pKa = 10.2HH75 pKa = 5.04QYY77 pKa = 10.7AAVKK81 pKa = 8.49YY82 pKa = 10.61QMMTVGGLYY91 pKa = 7.79QTNVKK96 pKa = 9.81QLSEE100 pKa = 4.58EE101 pKa = 3.93IEE103 pKa = 3.89KK104 pKa = 10.16TIIRR108 pKa = 11.84KK109 pKa = 9.13PDD111 pKa = 3.41LRR113 pKa = 11.84PTLVPNGDD121 pKa = 4.36SIDD124 pKa = 4.21IIPVNSMHH132 pKa = 6.25SHH134 pKa = 6.55PASAQFRR141 pKa = 11.84SSANSYY147 pKa = 11.33LNDD150 pKa = 3.53CVVRR154 pKa = 11.84AGYY157 pKa = 9.99RR158 pKa = 11.84PYY160 pKa = 10.68NISKK164 pKa = 9.92SRR166 pKa = 11.84RR167 pKa = 11.84DD168 pKa = 3.3IEE170 pKa = 4.35DD171 pKa = 3.32GCRR174 pKa = 11.84YY175 pKa = 9.88FYY177 pKa = 10.64HH178 pKa = 7.18SKK180 pKa = 10.69DD181 pKa = 3.61LSIKK185 pKa = 9.95YY186 pKa = 10.62SNDD189 pKa = 3.28KK190 pKa = 11.0IEE192 pKa = 4.55DD193 pKa = 3.33QSVFIMTDD201 pKa = 2.15VDD203 pKa = 4.4YY204 pKa = 11.26YY205 pKa = 11.63VDD207 pKa = 3.46MPRR210 pKa = 11.84WLKK213 pKa = 10.38LFRR216 pKa = 11.84PIVMYY221 pKa = 9.65TLSPTKK227 pKa = 10.45LSCSGPNDD235 pKa = 3.3KK236 pKa = 11.17AEE238 pKa = 3.73YY239 pKa = 10.16RR240 pKa = 11.84FNIDD244 pKa = 3.05GNVVHH249 pKa = 5.97YY250 pKa = 9.81HH251 pKa = 4.66VSGGGEE257 pKa = 3.98YY258 pKa = 8.71THH260 pKa = 6.48QLWNYY265 pKa = 8.41TGDD268 pKa = 3.41TVTCVDD274 pKa = 4.64DD275 pKa = 5.6DD276 pKa = 4.96GNLLTFDD283 pKa = 3.4IEE285 pKa = 4.17QRR287 pKa = 11.84KK288 pKa = 9.09VKK290 pKa = 10.46GDD292 pKa = 3.48EE293 pKa = 3.68QHH295 pKa = 6.71RR296 pKa = 11.84LIWLLPRR303 pKa = 11.84SKK305 pKa = 9.45VTNPLWHH312 pKa = 6.53YY313 pKa = 11.1LRR315 pKa = 11.84LDD317 pKa = 3.41WEE319 pKa = 4.45NNLLKK324 pKa = 10.69RR325 pKa = 11.84KK326 pKa = 10.35DD327 pKa = 3.44MTLKK331 pKa = 10.5GFNYY335 pKa = 10.05LLEE338 pKa = 5.11PIEE341 pKa = 5.06DD342 pKa = 3.77NLSIGLIGTKK352 pKa = 10.29YY353 pKa = 10.44SVEE356 pKa = 4.14IPGKK360 pKa = 9.29LYY362 pKa = 9.55EE363 pKa = 4.73AIRR366 pKa = 11.84IRR368 pKa = 11.84LAEE371 pKa = 4.08KK372 pKa = 9.39EE373 pKa = 4.13ATVFVSDD380 pKa = 3.63VEE382 pKa = 4.6RR383 pKa = 11.84MLKK386 pKa = 9.04EE387 pKa = 4.35AKK389 pKa = 9.19HH390 pKa = 4.97QSYY393 pKa = 7.97VTDD396 pKa = 3.7APILFKK402 pKa = 10.99CFAVDD407 pKa = 3.89VVFDD411 pKa = 4.15KK412 pKa = 11.55NVVKK416 pKa = 10.09TSNFAVTYY424 pKa = 9.0QAIPRR429 pKa = 11.84HH430 pKa = 5.69GALSTEE436 pKa = 4.67DD437 pKa = 3.87GNNPGQTTTSPLTSNPALFASKK459 pKa = 10.56GYY461 pKa = 9.96NADD464 pKa = 3.52KK465 pKa = 11.05ACIEE469 pKa = 4.0GRR471 pKa = 11.84IHH473 pKa = 7.31KK474 pKa = 9.31VANNKK479 pKa = 8.84KK480 pKa = 8.52WSKK483 pKa = 10.36KK484 pKa = 6.7YY485 pKa = 10.1HH486 pKa = 6.39DD487 pKa = 4.1YY488 pKa = 11.25ARR490 pKa = 11.84EE491 pKa = 4.21FIDD494 pKa = 3.57RR495 pKa = 11.84LVPKK499 pKa = 9.76RR500 pKa = 11.84YY501 pKa = 9.68RR502 pKa = 11.84GTGTPLSIGEE512 pKa = 3.83VDD514 pKa = 3.64LRR516 pKa = 11.84QEE518 pKa = 3.99KK519 pKa = 9.2VAQRR523 pKa = 11.84GRR525 pKa = 11.84FKK527 pKa = 10.7QVAPMMSLDD536 pKa = 3.6AGNAIKK542 pKa = 10.75AFIKK546 pKa = 10.04TEE548 pKa = 4.23TYY550 pKa = 11.13ASAKK554 pKa = 8.85PPRR557 pKa = 11.84NISTMSPEE565 pKa = 3.97LTIQMSAYY573 pKa = 8.67TLVFANILKK582 pKa = 8.5THH584 pKa = 6.9DD585 pKa = 3.56WYY587 pKa = 11.54CPGKK591 pKa = 10.22KK592 pKa = 9.59PRR594 pKa = 11.84DD595 pKa = 3.49IIKK598 pKa = 10.53RR599 pKa = 11.84LAHH602 pKa = 5.07VMKK605 pKa = 10.73AEE607 pKa = 3.94EE608 pKa = 4.42TEE610 pKa = 4.08DD611 pKa = 3.96VEE613 pKa = 4.9EE614 pKa = 5.2GDD616 pKa = 4.15YY617 pKa = 11.08TCLDD621 pKa = 3.59GTQSEE626 pKa = 5.13EE627 pKa = 4.01YY628 pKa = 10.56ARR630 pKa = 11.84NLLLPAYY637 pKa = 7.78MQYY640 pKa = 10.98LAPEE644 pKa = 4.34HH645 pKa = 6.0RR646 pKa = 11.84TMFKK650 pKa = 10.78DD651 pKa = 4.22LYY653 pKa = 10.04KK654 pKa = 10.46KK655 pKa = 10.45VYY657 pKa = 8.61KK658 pKa = 10.29QHH660 pKa = 6.28ATTSTGVSYY669 pKa = 11.1DD670 pKa = 2.81PGYY673 pKa = 8.72TVRR676 pKa = 11.84SGSPITTQAGTLANAFNVYY695 pKa = 10.06SALRR699 pKa = 11.84NMGYY703 pKa = 10.15VPDD706 pKa = 4.46EE707 pKa = 4.36AFDD710 pKa = 3.84MIGAIFGDD718 pKa = 4.47DD719 pKa = 3.57SCNPNYY725 pKa = 10.15KK726 pKa = 10.39GQFHH730 pKa = 7.0LFIEE734 pKa = 4.74QVAKK738 pKa = 10.41DD739 pKa = 3.55LGMIYY744 pKa = 10.34KK745 pKa = 10.57SNLRR749 pKa = 11.84PRR751 pKa = 11.84GEE753 pKa = 3.9PLLFLGRR760 pKa = 11.84YY761 pKa = 8.42FVDD764 pKa = 4.86PPTTDD769 pKa = 4.07DD770 pKa = 4.5SYY772 pKa = 12.1ADD774 pKa = 3.72PLRR777 pKa = 11.84TIGKK781 pKa = 8.69LHH783 pKa = 6.91ASTNKK788 pKa = 10.22SVTKK792 pKa = 10.09EE793 pKa = 3.26QAAANKK799 pKa = 9.98ALGYY803 pKa = 10.55LSTDD807 pKa = 2.8RR808 pKa = 11.84LTPIIGTWAARR819 pKa = 11.84VIQLTKK825 pKa = 9.98IRR827 pKa = 11.84TMKK830 pKa = 10.64GGTGEE835 pKa = 4.06EE836 pKa = 4.31QYY838 pKa = 10.79KK839 pKa = 10.71CSNAWPQKK847 pKa = 9.55DD848 pKa = 3.27EE849 pKa = 4.05TRR851 pKa = 11.84IRR853 pKa = 11.84EE854 pKa = 4.26CMATTLGITTAEE866 pKa = 4.74LIEE869 pKa = 4.53LDD871 pKa = 3.52KK872 pKa = 10.96KK873 pKa = 10.5IRR875 pKa = 11.84SVDD878 pKa = 3.65GLDD881 pKa = 3.45HH882 pKa = 6.71FPVIFDD888 pKa = 3.39TAYY891 pKa = 9.9KK892 pKa = 7.8HH893 pKa = 5.02TQVAVVDD900 pKa = 4.11GDD902 pKa = 4.16LVCTDD907 pKa = 3.42LHH909 pKa = 7.72QDD911 pKa = 3.21IEE913 pKa = 4.78PIHH916 pKa = 7.06DD917 pKa = 3.59EE918 pKa = 4.42SEE920 pKa = 4.3PKK922 pKa = 10.43ASCSGIQSPSSSHH935 pKa = 6.3RR936 pKa = 11.84SCSAKK941 pKa = 9.48ATEE944 pKa = 4.16RR945 pKa = 11.84PKK947 pKa = 10.76PGFTEE952 pKa = 4.84RR953 pKa = 11.84SFKK956 pKa = 10.3TSGRR960 pKa = 11.84RR961 pKa = 11.84ATGKK965 pKa = 9.54GHH967 pKa = 7.1PNRR970 pKa = 11.84IRR972 pKa = 11.84PNTRR976 pKa = 11.84KK977 pKa = 10.04SNRR980 pKa = 11.84VDD982 pKa = 3.26PRR984 pKa = 11.84DD985 pKa = 3.75DD986 pKa = 3.44VPRR989 pKa = 11.84TDD991 pKa = 3.32GRR993 pKa = 11.84RR994 pKa = 11.84INN996 pKa = 3.54
Molecular weight: 113.0 kDa
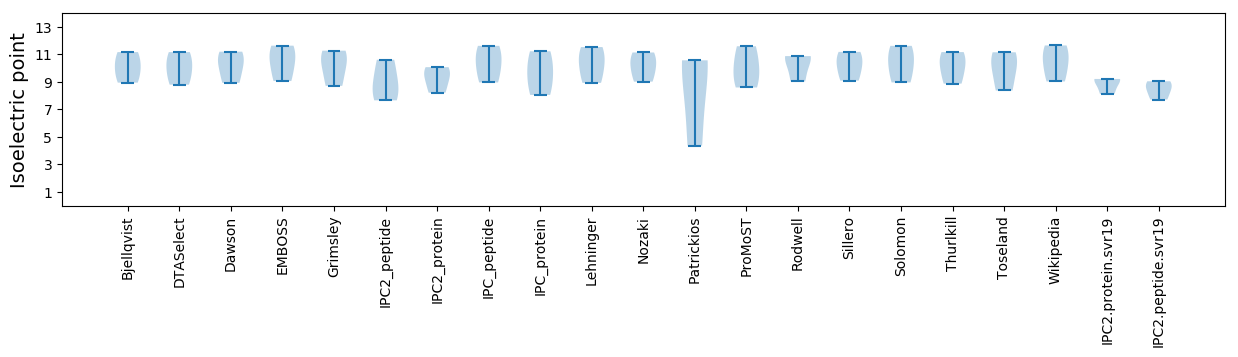
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGY7|A0A1L3KGY7_9VIRU Capsid protein OS=Wuhan house centipede virus 6 OX=1923710 PE=3 SV=1
MM1 pKa = 7.38VNKK4 pKa = 10.16NNQKK8 pKa = 10.18ARR10 pKa = 11.84NGKK13 pKa = 6.48TKK15 pKa = 10.49RR16 pKa = 11.84KK17 pKa = 7.1TRR19 pKa = 11.84NGQLQATRR27 pKa = 11.84EE28 pKa = 4.5GPASSRR34 pKa = 11.84SGMRR38 pKa = 11.84TGIPRR43 pKa = 11.84IRR45 pKa = 11.84NRR47 pKa = 11.84DD48 pKa = 3.36PYY50 pKa = 10.98SIEE53 pKa = 3.86VCNTEE58 pKa = 3.2IAMTITGTAAGGIIPAGGNIKK79 pKa = 9.9IMRR82 pKa = 11.84FDD84 pKa = 4.57DD85 pKa = 4.04SSTGNALNNKK95 pKa = 8.67HH96 pKa = 6.63WITKK100 pKa = 10.21LGLAYY105 pKa = 10.31DD106 pKa = 3.63KK107 pKa = 10.98FVIEE111 pKa = 4.23EE112 pKa = 4.25LSMKK116 pKa = 9.68FVPSLPFTAAGMSAMYY132 pKa = 10.15FDD134 pKa = 5.44SDD136 pKa = 4.01PSRR139 pKa = 11.84TTPPTSVASVSGDD152 pKa = 2.9MRR154 pKa = 11.84AVSKK158 pKa = 10.47QIYY161 pKa = 10.22AEE163 pKa = 4.41LPLKK167 pKa = 10.18VLRR170 pKa = 11.84NQLNRR175 pKa = 11.84LPQYY179 pKa = 7.97EE180 pKa = 4.42TFPGSGDD187 pKa = 3.31TGVATVGSINFVHH200 pKa = 7.51DD201 pKa = 4.93SIAMPNASTTGNITIGNVWMTYY223 pKa = 9.99RR224 pKa = 11.84IRR226 pKa = 11.84LINPSNAVAA235 pKa = 4.28
MM1 pKa = 7.38VNKK4 pKa = 10.16NNQKK8 pKa = 10.18ARR10 pKa = 11.84NGKK13 pKa = 6.48TKK15 pKa = 10.49RR16 pKa = 11.84KK17 pKa = 7.1TRR19 pKa = 11.84NGQLQATRR27 pKa = 11.84EE28 pKa = 4.5GPASSRR34 pKa = 11.84SGMRR38 pKa = 11.84TGIPRR43 pKa = 11.84IRR45 pKa = 11.84NRR47 pKa = 11.84DD48 pKa = 3.36PYY50 pKa = 10.98SIEE53 pKa = 3.86VCNTEE58 pKa = 3.2IAMTITGTAAGGIIPAGGNIKK79 pKa = 9.9IMRR82 pKa = 11.84FDD84 pKa = 4.57DD85 pKa = 4.04SSTGNALNNKK95 pKa = 8.67HH96 pKa = 6.63WITKK100 pKa = 10.21LGLAYY105 pKa = 10.31DD106 pKa = 3.63KK107 pKa = 10.98FVIEE111 pKa = 4.23EE112 pKa = 4.25LSMKK116 pKa = 9.68FVPSLPFTAAGMSAMYY132 pKa = 10.15FDD134 pKa = 5.44SDD136 pKa = 4.01PSRR139 pKa = 11.84TTPPTSVASVSGDD152 pKa = 2.9MRR154 pKa = 11.84AVSKK158 pKa = 10.47QIYY161 pKa = 10.22AEE163 pKa = 4.41LPLKK167 pKa = 10.18VLRR170 pKa = 11.84NQLNRR175 pKa = 11.84LPQYY179 pKa = 7.97EE180 pKa = 4.42TFPGSGDD187 pKa = 3.31TGVATVGSINFVHH200 pKa = 7.51DD201 pKa = 4.93SIAMPNASTTGNITIGNVWMTYY223 pKa = 9.99RR224 pKa = 11.84IRR226 pKa = 11.84LINPSNAVAA235 pKa = 4.28
Molecular weight: 25.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1387 |
156 |
996 |
462.3 |
51.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.705 ± 0.677 | 1.154 ± 0.467 |
5.84 ± 0.944 | 4.398 ± 0.959 |
2.812 ± 0.702 | 6.273 ± 0.875 |
2.235 ± 0.653 | 5.984 ± 0.614 |
6.2 ± 1.607 | 7.282 ± 0.797 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.623 | 5.696 ± 1.303 |
5.552 ± 0.308 | 3.317 ± 0.348 |
6.921 ± 1.333 | 6.921 ± 0.625 |
8.435 ± 1.36 | 6.273 ± 0.385 |
0.937 ± 0.092 | 4.47 ± 0.805 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
