
Stemphylium lycopersici mycovirus
Taxonomy: Viruses; Riboviria; unclassified Riboviria
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
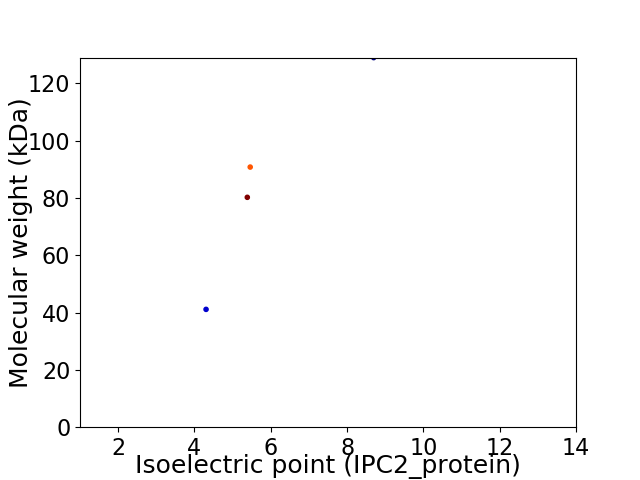
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346TJG4|A0A346TJG4_9VIRU Uncharacterized protein OS=Stemphylium lycopersici mycovirus OX=2499625 PE=4 SV=1
MM1 pKa = 7.32FCDD4 pKa = 4.81FEE6 pKa = 5.92PDD8 pKa = 3.45FPVFIPFLLRR18 pKa = 11.84NVEE21 pKa = 4.12SRR23 pKa = 11.84VFSAPALLTAQASGGFGGPKK43 pKa = 9.44SVRR46 pKa = 11.84VSIAHH51 pKa = 6.49FPLAAPDD58 pKa = 3.8AEE60 pKa = 4.08ALEE63 pKa = 4.74EE64 pKa = 4.28EE65 pKa = 4.98SCSGTAGQDD74 pKa = 3.39VFPVGVKK81 pKa = 8.99STVWCGGAVCSGSCGFDD98 pKa = 3.51GSCGISDD105 pKa = 3.23VDD107 pKa = 3.87EE108 pKa = 5.02RR109 pKa = 11.84GEE111 pKa = 4.5GYY113 pKa = 9.91FASTSFSSLVSVALAEE129 pKa = 4.0LGYY132 pKa = 10.71LGEE135 pKa = 4.26LCRR138 pKa = 11.84VQLSVGGMRR147 pKa = 11.84LDD149 pKa = 3.33ADD151 pKa = 3.81FRR153 pKa = 11.84GGARR157 pKa = 11.84PQTTVFDD164 pKa = 4.25AFAAEE169 pKa = 4.46AFFPPSLEE177 pKa = 3.96EE178 pKa = 4.04VVVEE182 pKa = 4.61DD183 pKa = 3.62VAPAKK188 pKa = 10.57VGAPADD194 pKa = 3.81IPGSMPLDD202 pKa = 3.54YY203 pKa = 10.8VVSLAEE209 pKa = 4.46TILPPAYY216 pKa = 10.51VEE218 pKa = 4.01AAKK221 pKa = 10.62ADD223 pKa = 3.81AVSLVPVSLPPPADD237 pKa = 3.72GPVLGVDD244 pKa = 4.19PGPAIPCVVNALFDD258 pKa = 3.98CAEE261 pKa = 4.45GVAQSTPGPVFAGDD275 pKa = 3.73CDD277 pKa = 4.33GEE279 pKa = 4.02IDD281 pKa = 4.07ARR283 pKa = 11.84LAVLQAGGTLPRR295 pKa = 11.84GCYY298 pKa = 10.3SIVGDD303 pKa = 4.24HH304 pKa = 5.95NWFNILRR311 pKa = 11.84GLARR315 pKa = 11.84WFASSGVSVRR325 pKa = 11.84GARR328 pKa = 11.84MDD330 pKa = 3.91LLLGGLSVARR340 pKa = 11.84DD341 pKa = 3.37DD342 pKa = 3.9RR343 pKa = 11.84LRR345 pKa = 11.84ALRR348 pKa = 11.84LLRR351 pKa = 11.84FLGLGRR357 pKa = 11.84GGDD360 pKa = 3.6EE361 pKa = 4.17AGVGDD366 pKa = 4.15RR367 pKa = 11.84VLFPPCARR375 pKa = 11.84AVVDD379 pKa = 4.19ARR381 pKa = 11.84LAYY384 pKa = 9.9LVWDD388 pKa = 4.74WIGGSSAADD397 pKa = 3.12
MM1 pKa = 7.32FCDD4 pKa = 4.81FEE6 pKa = 5.92PDD8 pKa = 3.45FPVFIPFLLRR18 pKa = 11.84NVEE21 pKa = 4.12SRR23 pKa = 11.84VFSAPALLTAQASGGFGGPKK43 pKa = 9.44SVRR46 pKa = 11.84VSIAHH51 pKa = 6.49FPLAAPDD58 pKa = 3.8AEE60 pKa = 4.08ALEE63 pKa = 4.74EE64 pKa = 4.28EE65 pKa = 4.98SCSGTAGQDD74 pKa = 3.39VFPVGVKK81 pKa = 8.99STVWCGGAVCSGSCGFDD98 pKa = 3.51GSCGISDD105 pKa = 3.23VDD107 pKa = 3.87EE108 pKa = 5.02RR109 pKa = 11.84GEE111 pKa = 4.5GYY113 pKa = 9.91FASTSFSSLVSVALAEE129 pKa = 4.0LGYY132 pKa = 10.71LGEE135 pKa = 4.26LCRR138 pKa = 11.84VQLSVGGMRR147 pKa = 11.84LDD149 pKa = 3.33ADD151 pKa = 3.81FRR153 pKa = 11.84GGARR157 pKa = 11.84PQTTVFDD164 pKa = 4.25AFAAEE169 pKa = 4.46AFFPPSLEE177 pKa = 3.96EE178 pKa = 4.04VVVEE182 pKa = 4.61DD183 pKa = 3.62VAPAKK188 pKa = 10.57VGAPADD194 pKa = 3.81IPGSMPLDD202 pKa = 3.54YY203 pKa = 10.8VVSLAEE209 pKa = 4.46TILPPAYY216 pKa = 10.51VEE218 pKa = 4.01AAKK221 pKa = 10.62ADD223 pKa = 3.81AVSLVPVSLPPPADD237 pKa = 3.72GPVLGVDD244 pKa = 4.19PGPAIPCVVNALFDD258 pKa = 3.98CAEE261 pKa = 4.45GVAQSTPGPVFAGDD275 pKa = 3.73CDD277 pKa = 4.33GEE279 pKa = 4.02IDD281 pKa = 4.07ARR283 pKa = 11.84LAVLQAGGTLPRR295 pKa = 11.84GCYY298 pKa = 10.3SIVGDD303 pKa = 4.24HH304 pKa = 5.95NWFNILRR311 pKa = 11.84GLARR315 pKa = 11.84WFASSGVSVRR325 pKa = 11.84GARR328 pKa = 11.84MDD330 pKa = 3.91LLLGGLSVARR340 pKa = 11.84DD341 pKa = 3.37DD342 pKa = 3.9RR343 pKa = 11.84LRR345 pKa = 11.84ALRR348 pKa = 11.84LLRR351 pKa = 11.84FLGLGRR357 pKa = 11.84GGDD360 pKa = 3.6EE361 pKa = 4.17AGVGDD366 pKa = 4.15RR367 pKa = 11.84VLFPPCARR375 pKa = 11.84AVVDD379 pKa = 4.19ARR381 pKa = 11.84LAYY384 pKa = 9.9LVWDD388 pKa = 4.74WIGGSSAADD397 pKa = 3.12
Molecular weight: 41.11 kDa
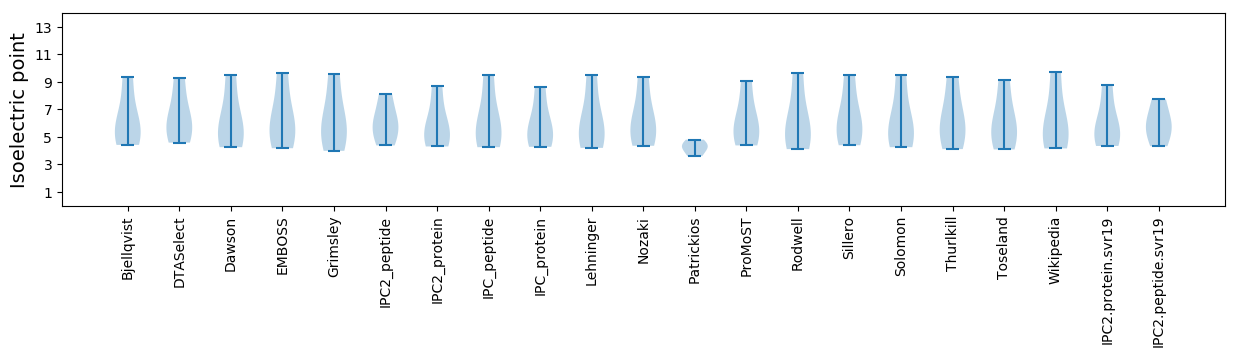
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346TJG2|A0A346TJG2_9VIRU Coat protein OS=Stemphylium lycopersici mycovirus OX=2499625 PE=4 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84SDD4 pKa = 4.32KK5 pKa = 11.17SLLLLGFRR13 pKa = 11.84NAGWLRR19 pKa = 11.84AQGEE23 pKa = 4.53HH24 pKa = 7.33PDD26 pKa = 3.74LGAIPKK32 pKa = 9.82RR33 pKa = 11.84AFVPEE38 pKa = 4.18PEE40 pKa = 4.66EE41 pKa = 4.21VDD43 pKa = 3.31ACLKK47 pKa = 10.66GIEE50 pKa = 4.9DD51 pKa = 3.7GWGDD55 pKa = 3.62GAFWCPTVIGSAFPVFFVLSNEE77 pKa = 3.84TRR79 pKa = 11.84NAFCHH84 pKa = 6.03SFSSVGGDD92 pKa = 3.66VNWLTFGLTDD102 pKa = 4.82ARR104 pKa = 11.84TLLYY108 pKa = 10.14RR109 pKa = 11.84RR110 pKa = 11.84PQVCVVAKK118 pKa = 9.45PRR120 pKa = 11.84PCVAYY125 pKa = 10.34HH126 pKa = 7.1RR127 pKa = 11.84DD128 pKa = 3.27LSSIARR134 pKa = 11.84VLRR137 pKa = 11.84YY138 pKa = 9.55GVLNSDD144 pKa = 4.59HH145 pKa = 6.98KK146 pKa = 11.5LCDD149 pKa = 3.33AYY151 pKa = 10.06TDD153 pKa = 4.46FVRR156 pKa = 11.84SPVARR161 pKa = 11.84TEE163 pKa = 4.35EE164 pKa = 3.94VWGTLGAYY172 pKa = 8.52PWGDD176 pKa = 3.84LDD178 pKa = 4.03TLVGQSTRR186 pKa = 11.84AEE188 pKa = 3.82YY189 pKa = 10.75DD190 pKa = 3.2AVKK193 pKa = 10.47DD194 pKa = 3.55AAVVGFLDD202 pKa = 4.04RR203 pKa = 11.84LWGKK207 pKa = 10.21KK208 pKa = 9.15RR209 pKa = 11.84GKK211 pKa = 9.74GAPRR215 pKa = 11.84DD216 pKa = 3.38RR217 pKa = 11.84KK218 pKa = 10.16VRR220 pKa = 11.84AKK222 pKa = 10.82AAMAWGDD229 pKa = 3.14ARR231 pKa = 11.84LWSRR235 pKa = 11.84EE236 pKa = 3.58VSFKK240 pKa = 10.33RR241 pKa = 11.84RR242 pKa = 11.84HH243 pKa = 5.67YY244 pKa = 10.58YY245 pKa = 10.41DD246 pKa = 3.03VATFLCVMDD255 pKa = 4.53EE256 pKa = 4.26VWGSRR261 pKa = 11.84DD262 pKa = 3.05VALRR266 pKa = 11.84GGATNAEE273 pKa = 4.37AGLQVGSFRR282 pKa = 11.84KK283 pKa = 9.29AVRR286 pKa = 11.84MVLSEE291 pKa = 4.42LVMGPASYY299 pKa = 10.63EE300 pKa = 3.8RR301 pKa = 11.84LYY303 pKa = 10.6LTIFWVFSTRR313 pKa = 11.84WWPSLVPVLIRR324 pKa = 11.84YY325 pKa = 7.18GTLGMSDD332 pKa = 4.34DD333 pKa = 4.98EE334 pKa = 4.24YY335 pKa = 10.98TAIHH339 pKa = 6.54KK340 pKa = 8.84EE341 pKa = 3.35ITAVVTSTWMVPGTCRR357 pKa = 11.84QHH359 pKa = 5.91QFSANFLNAEE369 pKa = 4.57DD370 pKa = 3.7LTGWSDD376 pKa = 3.47RR377 pKa = 11.84DD378 pKa = 3.67SLKK381 pKa = 11.22GGVCTEE387 pKa = 4.36IIKK390 pKa = 10.32FALATFEE397 pKa = 3.79YY398 pKa = 10.21RR399 pKa = 11.84ADD401 pKa = 3.47VRR403 pKa = 11.84GGGGEE408 pKa = 3.66RR409 pKa = 11.84GMPIIAKK416 pKa = 9.13EE417 pKa = 4.15GSPSEE422 pKa = 3.92YY423 pKa = 9.89LSRR426 pKa = 11.84YY427 pKa = 8.1RR428 pKa = 11.84AAMDD432 pKa = 3.2QLLRR436 pKa = 11.84PMYY439 pKa = 10.54AKK441 pKa = 10.69YY442 pKa = 9.87SANMTDD448 pKa = 4.11LRR450 pKa = 11.84SHH452 pKa = 5.46MEE454 pKa = 4.04KK455 pKa = 10.46RR456 pKa = 11.84MAWMSGGAVGRR467 pKa = 11.84RR468 pKa = 11.84AKK470 pKa = 10.32EE471 pKa = 3.52LLGPGIAPPGSSKK484 pKa = 11.08AYY486 pKa = 8.05VASRR490 pKa = 11.84ADD492 pKa = 3.6ADD494 pKa = 3.57QLTRR498 pKa = 11.84DD499 pKa = 3.45WGEE502 pKa = 3.46MRR504 pKa = 11.84IEE506 pKa = 3.98VGGKK510 pKa = 8.57GNEE513 pKa = 4.03RR514 pKa = 11.84GSEE517 pKa = 3.96RR518 pKa = 11.84TLLATDD524 pKa = 4.29LRR526 pKa = 11.84DD527 pKa = 3.66QVSEE531 pKa = 4.39SYY533 pKa = 10.83LLHH536 pKa = 6.56AFKK539 pKa = 10.77NRR541 pKa = 11.84YY542 pKa = 8.45GLIGVDD548 pKa = 3.33VGEE551 pKa = 4.82TPVEE555 pKa = 3.84MFKK558 pKa = 10.72RR559 pKa = 11.84HH560 pKa = 4.39VTVASATDD568 pKa = 3.54VPMSFHH574 pKa = 6.04GRR576 pKa = 11.84EE577 pKa = 4.17KK578 pKa = 10.67KK579 pKa = 10.54VLAAWDD585 pKa = 3.71YY586 pKa = 11.34SKK588 pKa = 10.67WDD590 pKa = 3.49HH591 pKa = 6.43HH592 pKa = 7.34VMLAEE597 pKa = 4.26RR598 pKa = 11.84LLLVQVMRR606 pKa = 11.84DD607 pKa = 3.31LVLEE611 pKa = 4.05YY612 pKa = 10.61VQRR615 pKa = 11.84DD616 pKa = 4.26DD617 pKa = 5.05IRR619 pKa = 11.84VDD621 pKa = 3.28MLKK624 pKa = 10.45EE625 pKa = 3.87LDD627 pKa = 3.77VLEE630 pKa = 4.61SSHH633 pKa = 6.51RR634 pKa = 11.84VAIYY638 pKa = 10.22RR639 pKa = 11.84SRR641 pKa = 11.84AYY643 pKa = 10.61ADD645 pKa = 3.36ARR647 pKa = 11.84YY648 pKa = 7.8TDD650 pKa = 3.58QVDD653 pKa = 3.02ALIRR657 pKa = 11.84DD658 pKa = 3.91GVKK661 pKa = 10.55AGFKK665 pKa = 9.59GQAVRR670 pKa = 11.84LSADD674 pKa = 3.22QVRR677 pKa = 11.84ITDD680 pKa = 3.53YY681 pKa = 11.29AGQQSGRR688 pKa = 11.84RR689 pKa = 11.84STLEE693 pKa = 3.6SNTFYY698 pKa = 11.23SRR700 pKa = 11.84ARR702 pKa = 11.84LLVRR706 pKa = 11.84DD707 pKa = 3.98AEE709 pKa = 4.25LAEE712 pKa = 4.55GEE714 pKa = 4.13RR715 pKa = 11.84SVYY718 pKa = 10.47LLNRR722 pKa = 11.84ADD724 pKa = 5.15DD725 pKa = 3.64VLEE728 pKa = 4.28VYY730 pKa = 10.17RR731 pKa = 11.84AWEE734 pKa = 3.88HH735 pKa = 5.7ARR737 pKa = 11.84NAIDD741 pKa = 4.24VMLLQGHH748 pKa = 6.65KK749 pKa = 10.49ANKK752 pKa = 8.77KK753 pKa = 7.94KK754 pKa = 10.5QVVQVRR760 pKa = 11.84TGVYY764 pKa = 10.06FRR766 pKa = 11.84ILYY769 pKa = 9.9AAGSMRR775 pKa = 11.84GFPPRR780 pKa = 11.84AVYY783 pKa = 10.5ACASAGPSMAASGGFDD799 pKa = 3.15PVEE802 pKa = 4.21RR803 pKa = 11.84LSSLSGALDD812 pKa = 3.24RR813 pKa = 11.84LARR816 pKa = 11.84RR817 pKa = 11.84GSGYY821 pKa = 8.88WVARR825 pKa = 11.84ALYY828 pKa = 10.44LEE830 pKa = 4.67AEE832 pKa = 4.39DD833 pKa = 4.46YY834 pKa = 10.92YY835 pKa = 11.5RR836 pKa = 11.84DD837 pKa = 3.5VRR839 pKa = 11.84VQLAVKK845 pKa = 9.87ADD847 pKa = 2.97KK848 pKa = 9.19FTRR851 pKa = 11.84FTIPRR856 pKa = 11.84EE857 pKa = 4.02VLRR860 pKa = 11.84ASPDD864 pKa = 3.31LGGCGVLPPGNYY876 pKa = 10.44DD877 pKa = 3.08FDD879 pKa = 5.82LSIKK883 pKa = 10.13CSEE886 pKa = 3.95IVGEE890 pKa = 5.54NGRR893 pKa = 11.84LWQAVRR899 pKa = 11.84EE900 pKa = 4.14RR901 pKa = 11.84LEE903 pKa = 3.86KK904 pKa = 9.38RR905 pKa = 11.84HH906 pKa = 5.91KK907 pKa = 10.51FRR909 pKa = 11.84GLADD913 pKa = 3.78LQGSAGRR920 pKa = 11.84AFSHH924 pKa = 7.05DD925 pKa = 3.53VPLDD929 pKa = 3.67VPEE932 pKa = 4.91KK933 pKa = 10.58AWQQWQKK940 pKa = 9.36RR941 pKa = 11.84WRR943 pKa = 11.84GDD945 pKa = 2.84RR946 pKa = 11.84AAQDD950 pKa = 3.45GRR952 pKa = 11.84GEE954 pKa = 4.18LLRR957 pKa = 11.84ARR959 pKa = 11.84LMGKK963 pKa = 8.43LWRR966 pKa = 11.84YY967 pKa = 7.77KK968 pKa = 10.43RR969 pKa = 11.84SWHH972 pKa = 6.93RR973 pKa = 11.84DD974 pKa = 3.06RR975 pKa = 11.84KK976 pKa = 9.24GDD978 pKa = 3.2AWGLEE983 pKa = 4.4GFGHH987 pKa = 7.59LLSLPVDD994 pKa = 4.3FFQLSWRR1001 pKa = 11.84ILRR1004 pKa = 11.84DD1005 pKa = 3.61CPGSGEE1011 pKa = 4.17VDD1013 pKa = 2.9RR1014 pKa = 11.84LMKK1017 pKa = 10.44GVQGPPAEE1025 pKa = 4.31GMLAKK1030 pKa = 10.37LWYY1033 pKa = 10.47GLGQDD1038 pKa = 4.1VISVLGRR1045 pKa = 11.84DD1046 pKa = 3.64VLEE1049 pKa = 4.88VIQSASAPGRR1059 pKa = 11.84EE1060 pKa = 4.98FAAEE1064 pKa = 4.12WGWLPLDD1071 pKa = 3.56VRR1073 pKa = 11.84EE1074 pKa = 4.15KK1075 pKa = 10.64FLRR1078 pKa = 11.84GHH1080 pKa = 7.52LGPAGAWMNLMPASFAPWLNQAVNFVLFQAFATGVRR1116 pKa = 11.84YY1117 pKa = 9.66HH1118 pKa = 6.69GSHH1121 pKa = 6.61WLLHH1125 pKa = 5.2VRR1127 pKa = 11.84QALTGAITRR1136 pKa = 11.84QFVDD1140 pKa = 3.27ARR1142 pKa = 11.84PLVVLHH1148 pKa = 6.72
MM1 pKa = 7.77RR2 pKa = 11.84SDD4 pKa = 4.32KK5 pKa = 11.17SLLLLGFRR13 pKa = 11.84NAGWLRR19 pKa = 11.84AQGEE23 pKa = 4.53HH24 pKa = 7.33PDD26 pKa = 3.74LGAIPKK32 pKa = 9.82RR33 pKa = 11.84AFVPEE38 pKa = 4.18PEE40 pKa = 4.66EE41 pKa = 4.21VDD43 pKa = 3.31ACLKK47 pKa = 10.66GIEE50 pKa = 4.9DD51 pKa = 3.7GWGDD55 pKa = 3.62GAFWCPTVIGSAFPVFFVLSNEE77 pKa = 3.84TRR79 pKa = 11.84NAFCHH84 pKa = 6.03SFSSVGGDD92 pKa = 3.66VNWLTFGLTDD102 pKa = 4.82ARR104 pKa = 11.84TLLYY108 pKa = 10.14RR109 pKa = 11.84RR110 pKa = 11.84PQVCVVAKK118 pKa = 9.45PRR120 pKa = 11.84PCVAYY125 pKa = 10.34HH126 pKa = 7.1RR127 pKa = 11.84DD128 pKa = 3.27LSSIARR134 pKa = 11.84VLRR137 pKa = 11.84YY138 pKa = 9.55GVLNSDD144 pKa = 4.59HH145 pKa = 6.98KK146 pKa = 11.5LCDD149 pKa = 3.33AYY151 pKa = 10.06TDD153 pKa = 4.46FVRR156 pKa = 11.84SPVARR161 pKa = 11.84TEE163 pKa = 4.35EE164 pKa = 3.94VWGTLGAYY172 pKa = 8.52PWGDD176 pKa = 3.84LDD178 pKa = 4.03TLVGQSTRR186 pKa = 11.84AEE188 pKa = 3.82YY189 pKa = 10.75DD190 pKa = 3.2AVKK193 pKa = 10.47DD194 pKa = 3.55AAVVGFLDD202 pKa = 4.04RR203 pKa = 11.84LWGKK207 pKa = 10.21KK208 pKa = 9.15RR209 pKa = 11.84GKK211 pKa = 9.74GAPRR215 pKa = 11.84DD216 pKa = 3.38RR217 pKa = 11.84KK218 pKa = 10.16VRR220 pKa = 11.84AKK222 pKa = 10.82AAMAWGDD229 pKa = 3.14ARR231 pKa = 11.84LWSRR235 pKa = 11.84EE236 pKa = 3.58VSFKK240 pKa = 10.33RR241 pKa = 11.84RR242 pKa = 11.84HH243 pKa = 5.67YY244 pKa = 10.58YY245 pKa = 10.41DD246 pKa = 3.03VATFLCVMDD255 pKa = 4.53EE256 pKa = 4.26VWGSRR261 pKa = 11.84DD262 pKa = 3.05VALRR266 pKa = 11.84GGATNAEE273 pKa = 4.37AGLQVGSFRR282 pKa = 11.84KK283 pKa = 9.29AVRR286 pKa = 11.84MVLSEE291 pKa = 4.42LVMGPASYY299 pKa = 10.63EE300 pKa = 3.8RR301 pKa = 11.84LYY303 pKa = 10.6LTIFWVFSTRR313 pKa = 11.84WWPSLVPVLIRR324 pKa = 11.84YY325 pKa = 7.18GTLGMSDD332 pKa = 4.34DD333 pKa = 4.98EE334 pKa = 4.24YY335 pKa = 10.98TAIHH339 pKa = 6.54KK340 pKa = 8.84EE341 pKa = 3.35ITAVVTSTWMVPGTCRR357 pKa = 11.84QHH359 pKa = 5.91QFSANFLNAEE369 pKa = 4.57DD370 pKa = 3.7LTGWSDD376 pKa = 3.47RR377 pKa = 11.84DD378 pKa = 3.67SLKK381 pKa = 11.22GGVCTEE387 pKa = 4.36IIKK390 pKa = 10.32FALATFEE397 pKa = 3.79YY398 pKa = 10.21RR399 pKa = 11.84ADD401 pKa = 3.47VRR403 pKa = 11.84GGGGEE408 pKa = 3.66RR409 pKa = 11.84GMPIIAKK416 pKa = 9.13EE417 pKa = 4.15GSPSEE422 pKa = 3.92YY423 pKa = 9.89LSRR426 pKa = 11.84YY427 pKa = 8.1RR428 pKa = 11.84AAMDD432 pKa = 3.2QLLRR436 pKa = 11.84PMYY439 pKa = 10.54AKK441 pKa = 10.69YY442 pKa = 9.87SANMTDD448 pKa = 4.11LRR450 pKa = 11.84SHH452 pKa = 5.46MEE454 pKa = 4.04KK455 pKa = 10.46RR456 pKa = 11.84MAWMSGGAVGRR467 pKa = 11.84RR468 pKa = 11.84AKK470 pKa = 10.32EE471 pKa = 3.52LLGPGIAPPGSSKK484 pKa = 11.08AYY486 pKa = 8.05VASRR490 pKa = 11.84ADD492 pKa = 3.6ADD494 pKa = 3.57QLTRR498 pKa = 11.84DD499 pKa = 3.45WGEE502 pKa = 3.46MRR504 pKa = 11.84IEE506 pKa = 3.98VGGKK510 pKa = 8.57GNEE513 pKa = 4.03RR514 pKa = 11.84GSEE517 pKa = 3.96RR518 pKa = 11.84TLLATDD524 pKa = 4.29LRR526 pKa = 11.84DD527 pKa = 3.66QVSEE531 pKa = 4.39SYY533 pKa = 10.83LLHH536 pKa = 6.56AFKK539 pKa = 10.77NRR541 pKa = 11.84YY542 pKa = 8.45GLIGVDD548 pKa = 3.33VGEE551 pKa = 4.82TPVEE555 pKa = 3.84MFKK558 pKa = 10.72RR559 pKa = 11.84HH560 pKa = 4.39VTVASATDD568 pKa = 3.54VPMSFHH574 pKa = 6.04GRR576 pKa = 11.84EE577 pKa = 4.17KK578 pKa = 10.67KK579 pKa = 10.54VLAAWDD585 pKa = 3.71YY586 pKa = 11.34SKK588 pKa = 10.67WDD590 pKa = 3.49HH591 pKa = 6.43HH592 pKa = 7.34VMLAEE597 pKa = 4.26RR598 pKa = 11.84LLLVQVMRR606 pKa = 11.84DD607 pKa = 3.31LVLEE611 pKa = 4.05YY612 pKa = 10.61VQRR615 pKa = 11.84DD616 pKa = 4.26DD617 pKa = 5.05IRR619 pKa = 11.84VDD621 pKa = 3.28MLKK624 pKa = 10.45EE625 pKa = 3.87LDD627 pKa = 3.77VLEE630 pKa = 4.61SSHH633 pKa = 6.51RR634 pKa = 11.84VAIYY638 pKa = 10.22RR639 pKa = 11.84SRR641 pKa = 11.84AYY643 pKa = 10.61ADD645 pKa = 3.36ARR647 pKa = 11.84YY648 pKa = 7.8TDD650 pKa = 3.58QVDD653 pKa = 3.02ALIRR657 pKa = 11.84DD658 pKa = 3.91GVKK661 pKa = 10.55AGFKK665 pKa = 9.59GQAVRR670 pKa = 11.84LSADD674 pKa = 3.22QVRR677 pKa = 11.84ITDD680 pKa = 3.53YY681 pKa = 11.29AGQQSGRR688 pKa = 11.84RR689 pKa = 11.84STLEE693 pKa = 3.6SNTFYY698 pKa = 11.23SRR700 pKa = 11.84ARR702 pKa = 11.84LLVRR706 pKa = 11.84DD707 pKa = 3.98AEE709 pKa = 4.25LAEE712 pKa = 4.55GEE714 pKa = 4.13RR715 pKa = 11.84SVYY718 pKa = 10.47LLNRR722 pKa = 11.84ADD724 pKa = 5.15DD725 pKa = 3.64VLEE728 pKa = 4.28VYY730 pKa = 10.17RR731 pKa = 11.84AWEE734 pKa = 3.88HH735 pKa = 5.7ARR737 pKa = 11.84NAIDD741 pKa = 4.24VMLLQGHH748 pKa = 6.65KK749 pKa = 10.49ANKK752 pKa = 8.77KK753 pKa = 7.94KK754 pKa = 10.5QVVQVRR760 pKa = 11.84TGVYY764 pKa = 10.06FRR766 pKa = 11.84ILYY769 pKa = 9.9AAGSMRR775 pKa = 11.84GFPPRR780 pKa = 11.84AVYY783 pKa = 10.5ACASAGPSMAASGGFDD799 pKa = 3.15PVEE802 pKa = 4.21RR803 pKa = 11.84LSSLSGALDD812 pKa = 3.24RR813 pKa = 11.84LARR816 pKa = 11.84RR817 pKa = 11.84GSGYY821 pKa = 8.88WVARR825 pKa = 11.84ALYY828 pKa = 10.44LEE830 pKa = 4.67AEE832 pKa = 4.39DD833 pKa = 4.46YY834 pKa = 10.92YY835 pKa = 11.5RR836 pKa = 11.84DD837 pKa = 3.5VRR839 pKa = 11.84VQLAVKK845 pKa = 9.87ADD847 pKa = 2.97KK848 pKa = 9.19FTRR851 pKa = 11.84FTIPRR856 pKa = 11.84EE857 pKa = 4.02VLRR860 pKa = 11.84ASPDD864 pKa = 3.31LGGCGVLPPGNYY876 pKa = 10.44DD877 pKa = 3.08FDD879 pKa = 5.82LSIKK883 pKa = 10.13CSEE886 pKa = 3.95IVGEE890 pKa = 5.54NGRR893 pKa = 11.84LWQAVRR899 pKa = 11.84EE900 pKa = 4.14RR901 pKa = 11.84LEE903 pKa = 3.86KK904 pKa = 9.38RR905 pKa = 11.84HH906 pKa = 5.91KK907 pKa = 10.51FRR909 pKa = 11.84GLADD913 pKa = 3.78LQGSAGRR920 pKa = 11.84AFSHH924 pKa = 7.05DD925 pKa = 3.53VPLDD929 pKa = 3.67VPEE932 pKa = 4.91KK933 pKa = 10.58AWQQWQKK940 pKa = 9.36RR941 pKa = 11.84WRR943 pKa = 11.84GDD945 pKa = 2.84RR946 pKa = 11.84AAQDD950 pKa = 3.45GRR952 pKa = 11.84GEE954 pKa = 4.18LLRR957 pKa = 11.84ARR959 pKa = 11.84LMGKK963 pKa = 8.43LWRR966 pKa = 11.84YY967 pKa = 7.77KK968 pKa = 10.43RR969 pKa = 11.84SWHH972 pKa = 6.93RR973 pKa = 11.84DD974 pKa = 3.06RR975 pKa = 11.84KK976 pKa = 9.24GDD978 pKa = 3.2AWGLEE983 pKa = 4.4GFGHH987 pKa = 7.59LLSLPVDD994 pKa = 4.3FFQLSWRR1001 pKa = 11.84ILRR1004 pKa = 11.84DD1005 pKa = 3.61CPGSGEE1011 pKa = 4.17VDD1013 pKa = 2.9RR1014 pKa = 11.84LMKK1017 pKa = 10.44GVQGPPAEE1025 pKa = 4.31GMLAKK1030 pKa = 10.37LWYY1033 pKa = 10.47GLGQDD1038 pKa = 4.1VISVLGRR1045 pKa = 11.84DD1046 pKa = 3.64VLEE1049 pKa = 4.88VIQSASAPGRR1059 pKa = 11.84EE1060 pKa = 4.98FAAEE1064 pKa = 4.12WGWLPLDD1071 pKa = 3.56VRR1073 pKa = 11.84EE1074 pKa = 4.15KK1075 pKa = 10.64FLRR1078 pKa = 11.84GHH1080 pKa = 7.52LGPAGAWMNLMPASFAPWLNQAVNFVLFQAFATGVRR1116 pKa = 11.84YY1117 pKa = 9.66HH1118 pKa = 6.69GSHH1121 pKa = 6.61WLLHH1125 pKa = 5.2VRR1127 pKa = 11.84QALTGAITRR1136 pKa = 11.84QFVDD1140 pKa = 3.27ARR1142 pKa = 11.84PLVVLHH1148 pKa = 6.72
Molecular weight: 128.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3143 |
397 |
1148 |
785.8 |
85.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.581 ± 0.836 | 1.336 ± 0.309 |
6.841 ± 0.178 | 5.154 ± 0.349 |
3.754 ± 0.401 | 11.04 ± 0.843 |
1.973 ± 0.272 | 2.195 ± 0.139 |
2.609 ± 0.689 | 9.8 ± 0.572 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.004 ± 0.229 | 1.75 ± 0.148 |
5.218 ± 0.824 | 2.195 ± 0.349 |
8.559 ± 0.649 | 6.109 ± 0.324 |
3.532 ± 0.248 | 9.131 ± 0.388 |
2.418 ± 0.31 | 2.8 ± 0.393 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
