
Blautia wexlerae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Blautia
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
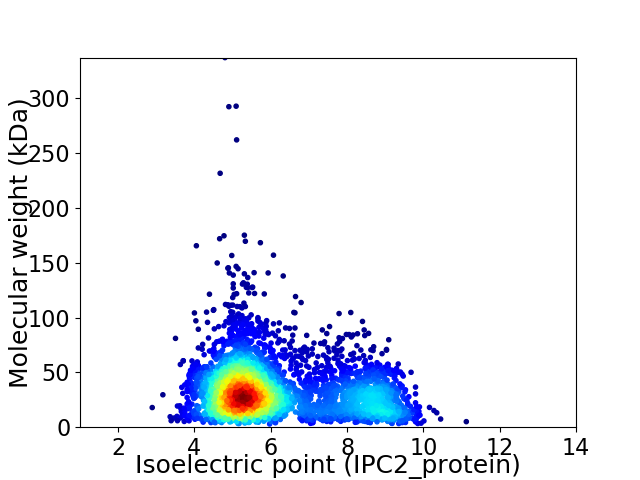
Virtual 2D-PAGE plot for 3715 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
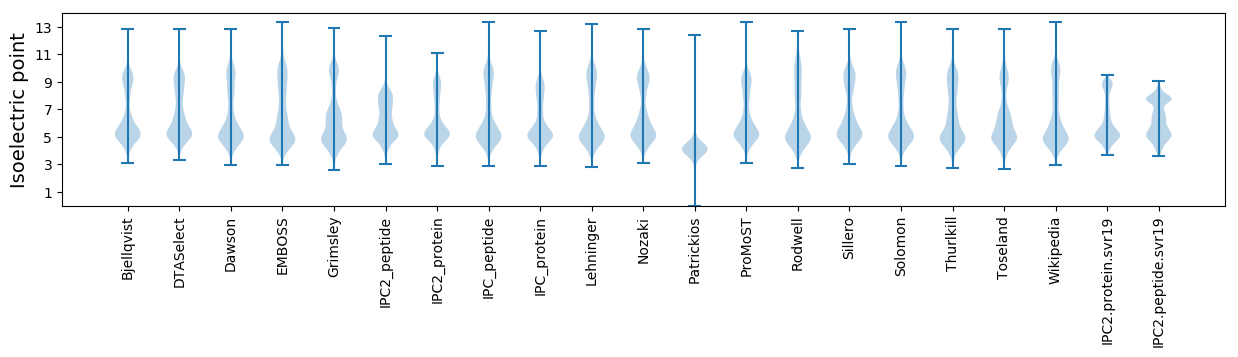
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A174C6S4|A0A174C6S4_9FIRM Cobalamin-binding protein OS=Blautia wexlerae OX=418240 GN=metH_2 PE=4 SV=1
MM1 pKa = 7.52KK2 pKa = 10.2NKK4 pKa = 10.44KK5 pKa = 10.03LMSILLVSAILGTTLGTTAVSAADD29 pKa = 3.79EE30 pKa = 4.55TSVSLWTWSPITRR43 pKa = 11.84TAEE46 pKa = 3.78KK47 pKa = 10.06MIDD50 pKa = 3.52AFEE53 pKa = 4.31KK54 pKa = 10.66ANPDD58 pKa = 2.84ITIDD62 pKa = 3.64YY63 pKa = 9.2TNYY66 pKa = 10.63NYY68 pKa = 10.64NPEE71 pKa = 4.0YY72 pKa = 10.71LQALSAASASDD83 pKa = 3.51NLADD87 pKa = 4.79IVGLQPGSLTQTYY100 pKa = 10.72SDD102 pKa = 4.02YY103 pKa = 11.44LIDD106 pKa = 4.42LSDD109 pKa = 3.6YY110 pKa = 11.6AKK112 pKa = 11.03AEE114 pKa = 4.12WGDD117 pKa = 3.78DD118 pKa = 3.16WTSVYY123 pKa = 11.46DD124 pKa = 4.95NVTQSQLQLGNKK136 pKa = 9.99DD137 pKa = 3.55GDD139 pKa = 3.95DD140 pKa = 3.34GHH142 pKa = 6.99YY143 pKa = 10.0ILPIEE148 pKa = 4.34TQDD151 pKa = 4.36IYY153 pKa = 11.87VEE155 pKa = 4.14YY156 pKa = 10.76NKK158 pKa = 9.78TLFEE162 pKa = 3.96QLGLKK167 pKa = 10.27VPTTYY172 pKa = 11.07DD173 pKa = 3.27EE174 pKa = 4.28LVEE177 pKa = 4.15VSKK180 pKa = 9.07TLRR183 pKa = 11.84DD184 pKa = 3.32NGYY187 pKa = 10.55APLFFGGADD196 pKa = 3.16GWQHH200 pKa = 5.97VNLLLMCTSQISDD213 pKa = 3.72TLFDD217 pKa = 3.5EE218 pKa = 4.86CQNGEE223 pKa = 4.25KK224 pKa = 10.31AWTCDD229 pKa = 3.1EE230 pKa = 4.51MKK232 pKa = 10.6QAMTNYY238 pKa = 10.04KK239 pKa = 10.25KK240 pKa = 10.92LFDD243 pKa = 5.32DD244 pKa = 5.77GVMQDD249 pKa = 4.09GSLSSTSYY257 pKa = 11.59SDD259 pKa = 3.15GTTLFLAGQAGMMLLGSWWAQEE281 pKa = 3.98YY282 pKa = 9.17TSEE285 pKa = 4.22DD286 pKa = 3.28VSDD289 pKa = 5.75AVANWDD295 pKa = 3.08YY296 pKa = 11.59DD297 pKa = 4.09YY298 pKa = 11.14FYY300 pKa = 11.36LPALEE305 pKa = 5.01EE306 pKa = 4.31GLSDD310 pKa = 3.46SKK312 pKa = 11.58AIGGVDD318 pKa = 3.98FGYY321 pKa = 11.07GITKK325 pKa = 10.14NCEE328 pKa = 3.88NPDD331 pKa = 4.2LAWKK335 pKa = 10.28ALVSFATGEE344 pKa = 4.26GAQEE348 pKa = 4.17IANDD352 pKa = 3.95MNNHH356 pKa = 6.32LSYY359 pKa = 11.23PNIEE363 pKa = 4.68PDD365 pKa = 3.04TSAMVEE371 pKa = 3.77RR372 pKa = 11.84DD373 pKa = 3.13GLQNVVDD380 pKa = 3.92EE381 pKa = 5.1FNRR384 pKa = 11.84SGKK387 pKa = 10.23DD388 pKa = 2.89IAAGLVNQRR397 pKa = 11.84IAEE400 pKa = 4.13PTIEE404 pKa = 4.11TAIQEE409 pKa = 4.18AMQGLIGGTYY419 pKa = 10.31SVDD422 pKa = 3.11EE423 pKa = 4.46ATQHH427 pKa = 6.12IQDD430 pKa = 4.38AQDD433 pKa = 3.15ALL435 pKa = 3.86
MM1 pKa = 7.52KK2 pKa = 10.2NKK4 pKa = 10.44KK5 pKa = 10.03LMSILLVSAILGTTLGTTAVSAADD29 pKa = 3.79EE30 pKa = 4.55TSVSLWTWSPITRR43 pKa = 11.84TAEE46 pKa = 3.78KK47 pKa = 10.06MIDD50 pKa = 3.52AFEE53 pKa = 4.31KK54 pKa = 10.66ANPDD58 pKa = 2.84ITIDD62 pKa = 3.64YY63 pKa = 9.2TNYY66 pKa = 10.63NYY68 pKa = 10.64NPEE71 pKa = 4.0YY72 pKa = 10.71LQALSAASASDD83 pKa = 3.51NLADD87 pKa = 4.79IVGLQPGSLTQTYY100 pKa = 10.72SDD102 pKa = 4.02YY103 pKa = 11.44LIDD106 pKa = 4.42LSDD109 pKa = 3.6YY110 pKa = 11.6AKK112 pKa = 11.03AEE114 pKa = 4.12WGDD117 pKa = 3.78DD118 pKa = 3.16WTSVYY123 pKa = 11.46DD124 pKa = 4.95NVTQSQLQLGNKK136 pKa = 9.99DD137 pKa = 3.55GDD139 pKa = 3.95DD140 pKa = 3.34GHH142 pKa = 6.99YY143 pKa = 10.0ILPIEE148 pKa = 4.34TQDD151 pKa = 4.36IYY153 pKa = 11.87VEE155 pKa = 4.14YY156 pKa = 10.76NKK158 pKa = 9.78TLFEE162 pKa = 3.96QLGLKK167 pKa = 10.27VPTTYY172 pKa = 11.07DD173 pKa = 3.27EE174 pKa = 4.28LVEE177 pKa = 4.15VSKK180 pKa = 9.07TLRR183 pKa = 11.84DD184 pKa = 3.32NGYY187 pKa = 10.55APLFFGGADD196 pKa = 3.16GWQHH200 pKa = 5.97VNLLLMCTSQISDD213 pKa = 3.72TLFDD217 pKa = 3.5EE218 pKa = 4.86CQNGEE223 pKa = 4.25KK224 pKa = 10.31AWTCDD229 pKa = 3.1EE230 pKa = 4.51MKK232 pKa = 10.6QAMTNYY238 pKa = 10.04KK239 pKa = 10.25KK240 pKa = 10.92LFDD243 pKa = 5.32DD244 pKa = 5.77GVMQDD249 pKa = 4.09GSLSSTSYY257 pKa = 11.59SDD259 pKa = 3.15GTTLFLAGQAGMMLLGSWWAQEE281 pKa = 3.98YY282 pKa = 9.17TSEE285 pKa = 4.22DD286 pKa = 3.28VSDD289 pKa = 5.75AVANWDD295 pKa = 3.08YY296 pKa = 11.59DD297 pKa = 4.09YY298 pKa = 11.14FYY300 pKa = 11.36LPALEE305 pKa = 5.01EE306 pKa = 4.31GLSDD310 pKa = 3.46SKK312 pKa = 11.58AIGGVDD318 pKa = 3.98FGYY321 pKa = 11.07GITKK325 pKa = 10.14NCEE328 pKa = 3.88NPDD331 pKa = 4.2LAWKK335 pKa = 10.28ALVSFATGEE344 pKa = 4.26GAQEE348 pKa = 4.17IANDD352 pKa = 3.95MNNHH356 pKa = 6.32LSYY359 pKa = 11.23PNIEE363 pKa = 4.68PDD365 pKa = 3.04TSAMVEE371 pKa = 3.77RR372 pKa = 11.84DD373 pKa = 3.13GLQNVVDD380 pKa = 3.92EE381 pKa = 5.1FNRR384 pKa = 11.84SGKK387 pKa = 10.23DD388 pKa = 2.89IAAGLVNQRR397 pKa = 11.84IAEE400 pKa = 4.13PTIEE404 pKa = 4.11TAIQEE409 pKa = 4.18AMQGLIGGTYY419 pKa = 10.31SVDD422 pKa = 3.11EE423 pKa = 4.46ATQHH427 pKa = 6.12IQDD430 pKa = 4.38AQDD433 pKa = 3.15ALL435 pKa = 3.86
Molecular weight: 47.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A174DFB4|A0A174DFB4_9FIRM Dihydroorotase OS=Blautia wexlerae OX=418240 GN=pyrC PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.41GRR39 pKa = 11.84KK40 pKa = 8.72HH41 pKa = 6.34LSAA44 pKa = 5.8
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.15GGRR28 pKa = 11.84KK29 pKa = 8.69VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.41GRR39 pKa = 11.84KK40 pKa = 8.72HH41 pKa = 6.34LSAA44 pKa = 5.8
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1165781 |
29 |
2994 |
313.8 |
35.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.211 ± 0.044 | 1.556 ± 0.018 |
5.773 ± 0.035 | 7.655 ± 0.045 |
4.017 ± 0.029 | 6.927 ± 0.042 |
1.769 ± 0.019 | 7.308 ± 0.036 |
7.226 ± 0.036 | 8.717 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.151 ± 0.023 | 4.466 ± 0.027 |
3.259 ± 0.022 | 3.399 ± 0.024 |
4.323 ± 0.036 | 5.775 ± 0.033 |
5.54 ± 0.035 | 6.756 ± 0.037 |
0.948 ± 0.013 | 4.222 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
