
Thermoplasmatales archaeon SCGC AB-540-F20
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Thermoplasmatales; unclassified Thermoplasmatales
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
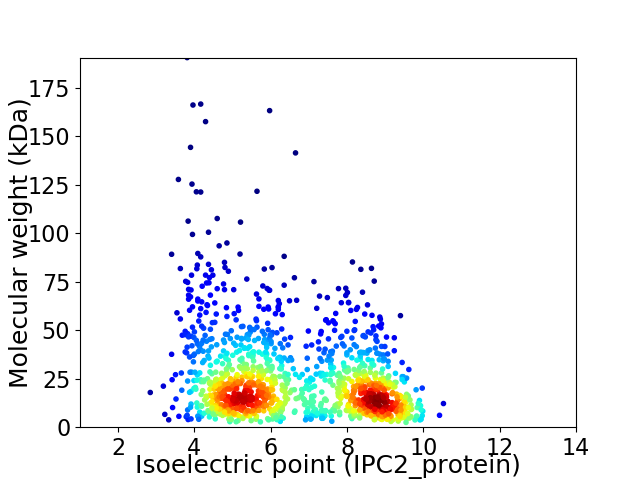
Virtual 2D-PAGE plot for 1255 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7TFT6|M7TFT6_9ARCH Thioredoxin OS=Thermoplasmatales archaeon SCGC AB-540-F20 OX=1242866 GN=MBGDF03_00437 PE=3 SV=1
MM1 pKa = 7.48KK2 pKa = 10.31KK3 pKa = 9.78IISSILVILFVLAGLEE19 pKa = 4.25VIGISNDD26 pKa = 2.89KK27 pKa = 10.17TIEE30 pKa = 3.92NFYY33 pKa = 11.09EE34 pKa = 4.3SDD36 pKa = 3.54FKK38 pKa = 11.72GLFDD42 pKa = 5.91DD43 pKa = 3.99ISVYY47 pKa = 10.16RR48 pKa = 11.84YY49 pKa = 7.52QTACQGKK56 pKa = 8.73VPFSMWSIDD65 pKa = 3.44ALYY68 pKa = 10.37TNSEE72 pKa = 4.17NVNEE76 pKa = 4.06NLYY79 pKa = 10.71FEE81 pKa = 4.97QIHH84 pKa = 6.67SIGFDD89 pKa = 2.99RR90 pKa = 11.84DD91 pKa = 3.64YY92 pKa = 11.16PIGEE96 pKa = 4.91IIWQYY101 pKa = 11.54LLTIYY106 pKa = 10.58DD107 pKa = 4.46PSPKK111 pKa = 10.08AIAPIEE117 pKa = 4.84DD118 pKa = 3.55INGDD122 pKa = 3.96GISDD126 pKa = 4.09VIVCSEE132 pKa = 4.44DD133 pKa = 3.12DD134 pKa = 3.36HH135 pKa = 7.27VRR137 pKa = 11.84CFGGGAIGTGVVLWGHH153 pKa = 6.73EE154 pKa = 3.99IYY156 pKa = 10.97AGDD159 pKa = 3.92VYY161 pKa = 11.12NQNGLDD167 pKa = 4.12IIDD170 pKa = 4.94DD171 pKa = 3.83VDD173 pKa = 4.14GDD175 pKa = 4.04GYY177 pKa = 11.31EE178 pKa = 4.14DD179 pKa = 3.99VVVGAAWGARR189 pKa = 11.84LIRR192 pKa = 11.84CISGIDD198 pKa = 3.7GSTIWTHH205 pKa = 5.82DD206 pKa = 3.06THH208 pKa = 7.03EE209 pKa = 4.42YY210 pKa = 10.96GGGGWVYY217 pKa = 9.96MVNCSYY223 pKa = 11.21DD224 pKa = 3.45YY225 pKa = 11.52DD226 pKa = 5.01GDD228 pKa = 4.14GVKK231 pKa = 10.51DD232 pKa = 3.7VLATCGDD239 pKa = 4.09DD240 pKa = 5.19SSDD243 pKa = 3.43TGPKK247 pKa = 9.28RR248 pKa = 11.84VYY250 pKa = 10.79CLDD253 pKa = 3.75GEE255 pKa = 4.74DD256 pKa = 5.15GVSIWEE262 pKa = 4.36RR263 pKa = 11.84PLGGPGFSVIGVEE276 pKa = 5.14DD277 pKa = 3.79FTGDD281 pKa = 4.31GIPDD285 pKa = 3.67VVAGCSNEE293 pKa = 3.91AEE295 pKa = 4.38TIGYY299 pKa = 10.05AKK301 pKa = 10.26GINGDD306 pKa = 3.04TGAQVWSKK314 pKa = 8.4TASGSSVWALEE325 pKa = 3.87QIEE328 pKa = 5.72DD329 pKa = 3.55ITGDD333 pKa = 3.73GIKK336 pKa = 10.5DD337 pKa = 3.52VIIGDD342 pKa = 3.95FSGNIYY348 pKa = 10.81GLDD351 pKa = 3.48ATDD354 pKa = 4.56GGQEE358 pKa = 3.95YY359 pKa = 8.42STSIGTAIITRR370 pKa = 11.84FAKK373 pKa = 10.82LNDD376 pKa = 3.28VDD378 pKa = 4.73YY379 pKa = 10.74SGHH382 pKa = 6.33PEE384 pKa = 3.61IVPAHH389 pKa = 6.11SSIHH393 pKa = 4.42TTQLIDD399 pKa = 4.25AEE401 pKa = 4.76DD402 pKa = 4.14GSIIWSHH409 pKa = 5.84GVADD413 pKa = 4.11QPWNVARR420 pKa = 11.84ISDD423 pKa = 3.78VSGDD427 pKa = 4.0GIDD430 pKa = 3.98DD431 pKa = 3.87VLVGTLYY438 pKa = 11.0NSNYY442 pKa = 10.12CYY444 pKa = 10.53FLDD447 pKa = 4.03GTNGSEE453 pKa = 5.01LEE455 pKa = 4.32TIAYY459 pKa = 7.64GQAVDD464 pKa = 5.56AINAIPDD471 pKa = 3.61VVADD475 pKa = 4.18GSMEE479 pKa = 3.98MVAGGRR485 pKa = 11.84DD486 pKa = 3.47GKK488 pKa = 10.3IVCFSGGLNAFSPNVEE504 pKa = 3.88VEE506 pKa = 4.03ADD508 pKa = 4.74FKK510 pKa = 11.86ADD512 pKa = 3.08ITEE515 pKa = 4.35GKK517 pKa = 10.62APLTVHH523 pKa = 6.25FTDD526 pKa = 5.58LSIAEE531 pKa = 4.28NTTITSWEE539 pKa = 3.87WDD541 pKa = 3.51FDD543 pKa = 3.76NDD545 pKa = 5.42DD546 pKa = 4.77IIDD549 pKa = 4.05SEE551 pKa = 4.39EE552 pKa = 3.91QNPVWTYY559 pKa = 9.9TEE561 pKa = 4.02GGIYY565 pKa = 9.26TVSLTVSDD573 pKa = 3.59GTIFDD578 pKa = 4.42TEE580 pKa = 4.25TKK582 pKa = 10.23ADD584 pKa = 4.2YY585 pKa = 10.58IAVSQITLEE594 pKa = 4.21IGDD597 pKa = 3.6ITGGLFNVNAVIKK610 pKa = 9.53NTGTIEE616 pKa = 3.73ATGAHH621 pKa = 5.12WRR623 pKa = 11.84ITLEE627 pKa = 3.96GGFILLGRR635 pKa = 11.84NTSGEE640 pKa = 4.16NLSITAGGEE649 pKa = 3.98EE650 pKa = 4.5TVSSKK655 pKa = 11.12LILGFGQTTVTVEE668 pKa = 3.32VWIPDD673 pKa = 3.71GPSDD677 pKa = 3.5TRR679 pKa = 11.84EE680 pKa = 3.52QGGFVIIFFIKK691 pKa = 10.22VNPGGII697 pKa = 3.57
MM1 pKa = 7.48KK2 pKa = 10.31KK3 pKa = 9.78IISSILVILFVLAGLEE19 pKa = 4.25VIGISNDD26 pKa = 2.89KK27 pKa = 10.17TIEE30 pKa = 3.92NFYY33 pKa = 11.09EE34 pKa = 4.3SDD36 pKa = 3.54FKK38 pKa = 11.72GLFDD42 pKa = 5.91DD43 pKa = 3.99ISVYY47 pKa = 10.16RR48 pKa = 11.84YY49 pKa = 7.52QTACQGKK56 pKa = 8.73VPFSMWSIDD65 pKa = 3.44ALYY68 pKa = 10.37TNSEE72 pKa = 4.17NVNEE76 pKa = 4.06NLYY79 pKa = 10.71FEE81 pKa = 4.97QIHH84 pKa = 6.67SIGFDD89 pKa = 2.99RR90 pKa = 11.84DD91 pKa = 3.64YY92 pKa = 11.16PIGEE96 pKa = 4.91IIWQYY101 pKa = 11.54LLTIYY106 pKa = 10.58DD107 pKa = 4.46PSPKK111 pKa = 10.08AIAPIEE117 pKa = 4.84DD118 pKa = 3.55INGDD122 pKa = 3.96GISDD126 pKa = 4.09VIVCSEE132 pKa = 4.44DD133 pKa = 3.12DD134 pKa = 3.36HH135 pKa = 7.27VRR137 pKa = 11.84CFGGGAIGTGVVLWGHH153 pKa = 6.73EE154 pKa = 3.99IYY156 pKa = 10.97AGDD159 pKa = 3.92VYY161 pKa = 11.12NQNGLDD167 pKa = 4.12IIDD170 pKa = 4.94DD171 pKa = 3.83VDD173 pKa = 4.14GDD175 pKa = 4.04GYY177 pKa = 11.31EE178 pKa = 4.14DD179 pKa = 3.99VVVGAAWGARR189 pKa = 11.84LIRR192 pKa = 11.84CISGIDD198 pKa = 3.7GSTIWTHH205 pKa = 5.82DD206 pKa = 3.06THH208 pKa = 7.03EE209 pKa = 4.42YY210 pKa = 10.96GGGGWVYY217 pKa = 9.96MVNCSYY223 pKa = 11.21DD224 pKa = 3.45YY225 pKa = 11.52DD226 pKa = 5.01GDD228 pKa = 4.14GVKK231 pKa = 10.51DD232 pKa = 3.7VLATCGDD239 pKa = 4.09DD240 pKa = 5.19SSDD243 pKa = 3.43TGPKK247 pKa = 9.28RR248 pKa = 11.84VYY250 pKa = 10.79CLDD253 pKa = 3.75GEE255 pKa = 4.74DD256 pKa = 5.15GVSIWEE262 pKa = 4.36RR263 pKa = 11.84PLGGPGFSVIGVEE276 pKa = 5.14DD277 pKa = 3.79FTGDD281 pKa = 4.31GIPDD285 pKa = 3.67VVAGCSNEE293 pKa = 3.91AEE295 pKa = 4.38TIGYY299 pKa = 10.05AKK301 pKa = 10.26GINGDD306 pKa = 3.04TGAQVWSKK314 pKa = 8.4TASGSSVWALEE325 pKa = 3.87QIEE328 pKa = 5.72DD329 pKa = 3.55ITGDD333 pKa = 3.73GIKK336 pKa = 10.5DD337 pKa = 3.52VIIGDD342 pKa = 3.95FSGNIYY348 pKa = 10.81GLDD351 pKa = 3.48ATDD354 pKa = 4.56GGQEE358 pKa = 3.95YY359 pKa = 8.42STSIGTAIITRR370 pKa = 11.84FAKK373 pKa = 10.82LNDD376 pKa = 3.28VDD378 pKa = 4.73YY379 pKa = 10.74SGHH382 pKa = 6.33PEE384 pKa = 3.61IVPAHH389 pKa = 6.11SSIHH393 pKa = 4.42TTQLIDD399 pKa = 4.25AEE401 pKa = 4.76DD402 pKa = 4.14GSIIWSHH409 pKa = 5.84GVADD413 pKa = 4.11QPWNVARR420 pKa = 11.84ISDD423 pKa = 3.78VSGDD427 pKa = 4.0GIDD430 pKa = 3.98DD431 pKa = 3.87VLVGTLYY438 pKa = 11.0NSNYY442 pKa = 10.12CYY444 pKa = 10.53FLDD447 pKa = 4.03GTNGSEE453 pKa = 5.01LEE455 pKa = 4.32TIAYY459 pKa = 7.64GQAVDD464 pKa = 5.56AINAIPDD471 pKa = 3.61VVADD475 pKa = 4.18GSMEE479 pKa = 3.98MVAGGRR485 pKa = 11.84DD486 pKa = 3.47GKK488 pKa = 10.3IVCFSGGLNAFSPNVEE504 pKa = 3.88VEE506 pKa = 4.03ADD508 pKa = 4.74FKK510 pKa = 11.86ADD512 pKa = 3.08ITEE515 pKa = 4.35GKK517 pKa = 10.62APLTVHH523 pKa = 6.25FTDD526 pKa = 5.58LSIAEE531 pKa = 4.28NTTITSWEE539 pKa = 3.87WDD541 pKa = 3.51FDD543 pKa = 3.76NDD545 pKa = 5.42DD546 pKa = 4.77IIDD549 pKa = 4.05SEE551 pKa = 4.39EE552 pKa = 3.91QNPVWTYY559 pKa = 9.9TEE561 pKa = 4.02GGIYY565 pKa = 9.26TVSLTVSDD573 pKa = 3.59GTIFDD578 pKa = 4.42TEE580 pKa = 4.25TKK582 pKa = 10.23ADD584 pKa = 4.2YY585 pKa = 10.58IAVSQITLEE594 pKa = 4.21IGDD597 pKa = 3.6ITGGLFNVNAVIKK610 pKa = 9.53NTGTIEE616 pKa = 3.73ATGAHH621 pKa = 5.12WRR623 pKa = 11.84ITLEE627 pKa = 3.96GGFILLGRR635 pKa = 11.84NTSGEE640 pKa = 4.16NLSITAGGEE649 pKa = 3.98EE650 pKa = 4.5TVSSKK655 pKa = 11.12LILGFGQTTVTVEE668 pKa = 3.32VWIPDD673 pKa = 3.71GPSDD677 pKa = 3.5TRR679 pKa = 11.84EE680 pKa = 3.52QGGFVIIFFIKK691 pKa = 10.22VNPGGII697 pKa = 3.57
Molecular weight: 74.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7TDA3|M7TDA3_9ARCH Uncharacterized protein OS=Thermoplasmatales archaeon SCGC AB-540-F20 OX=1242866 GN=MBGDF03_01192 PE=4 SV=1
SS1 pKa = 7.15IYY3 pKa = 10.64NFCFHH8 pKa = 7.01SIIVLSRR15 pKa = 11.84NYY17 pKa = 9.81FLRR20 pKa = 11.84VNKK23 pKa = 8.87VTDD26 pKa = 3.81LVYY29 pKa = 10.13MKK31 pKa = 10.62KK32 pKa = 10.52LNQKK36 pKa = 8.11KK37 pKa = 9.05VRR39 pKa = 11.84WIVRR43 pKa = 11.84EE44 pKa = 3.82MEE46 pKa = 4.1KK47 pKa = 10.72GDD49 pKa = 3.35RR50 pKa = 11.84SVYY53 pKa = 10.33RR54 pKa = 11.84IAKK57 pKa = 8.92TMDD60 pKa = 2.63ITPRR64 pKa = 11.84WVRR67 pKa = 11.84EE68 pKa = 3.71IYY70 pKa = 9.96RR71 pKa = 11.84DD72 pKa = 3.51HH73 pKa = 6.52QRR75 pKa = 11.84TGRR78 pKa = 11.84YY79 pKa = 8.96LYY81 pKa = 7.1PTKK84 pKa = 10.27PGRR87 pKa = 11.84KK88 pKa = 8.04PRR90 pKa = 11.84SISNEE95 pKa = 3.8RR96 pKa = 11.84EE97 pKa = 3.84RR98 pKa = 11.84SCC100 pKa = 5.17
SS1 pKa = 7.15IYY3 pKa = 10.64NFCFHH8 pKa = 7.01SIIVLSRR15 pKa = 11.84NYY17 pKa = 9.81FLRR20 pKa = 11.84VNKK23 pKa = 8.87VTDD26 pKa = 3.81LVYY29 pKa = 10.13MKK31 pKa = 10.62KK32 pKa = 10.52LNQKK36 pKa = 8.11KK37 pKa = 9.05VRR39 pKa = 11.84WIVRR43 pKa = 11.84EE44 pKa = 3.82MEE46 pKa = 4.1KK47 pKa = 10.72GDD49 pKa = 3.35RR50 pKa = 11.84SVYY53 pKa = 10.33RR54 pKa = 11.84IAKK57 pKa = 8.92TMDD60 pKa = 2.63ITPRR64 pKa = 11.84WVRR67 pKa = 11.84EE68 pKa = 3.71IYY70 pKa = 9.96RR71 pKa = 11.84DD72 pKa = 3.51HH73 pKa = 6.52QRR75 pKa = 11.84TGRR78 pKa = 11.84YY79 pKa = 8.96LYY81 pKa = 7.1PTKK84 pKa = 10.27PGRR87 pKa = 11.84KK88 pKa = 8.04PRR90 pKa = 11.84SISNEE95 pKa = 3.8RR96 pKa = 11.84EE97 pKa = 3.84RR98 pKa = 11.84SCC100 pKa = 5.17
Molecular weight: 12.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
295448 |
22 |
1733 |
235.4 |
26.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.334 ± 0.068 | 1.359 ± 0.038 |
6.193 ± 0.072 | 6.733 ± 0.072 |
4.581 ± 0.055 | 7.009 ± 0.068 |
1.842 ± 0.031 | 9.104 ± 0.094 |
7.555 ± 0.134 | 8.35 ± 0.079 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.391 ± 0.037 | 5.597 ± 0.089 |
3.886 ± 0.051 | 2.447 ± 0.035 |
3.692 ± 0.061 | 6.453 ± 0.083 |
5.702 ± 0.075 | 6.26 ± 0.054 |
1.465 ± 0.042 | 4.047 ± 0.056 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
