
Muribaculaceae bacterium Z82
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Muribaculaceae; unclassified Muribaculaceae
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
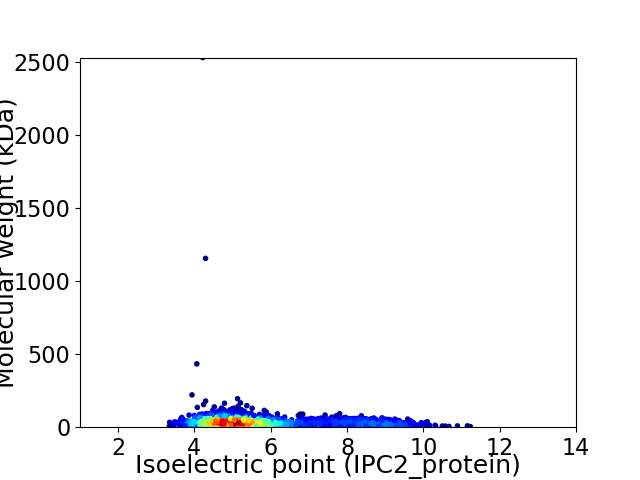
Virtual 2D-PAGE plot for 2214 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7C9JDP7|A0A7C9JDP7_9BACT Uncharacterized protein OS=Muribaculaceae bacterium Z82 OX=2304548 GN=D1639_06380 PE=4 SV=1
MM1 pKa = 7.51ALSLLVLGACSAPAPTDD18 pKa = 3.45NSEE21 pKa = 4.0KK22 pKa = 10.58AAPEE26 pKa = 4.08TPAAPLPASDD36 pKa = 4.59VMKK39 pKa = 10.88NDD41 pKa = 5.13DD42 pKa = 3.68PTQTSGFFAFDD53 pKa = 3.05SALFQGKK60 pKa = 7.86LTGCSYY66 pKa = 11.28AQAGTLLVCADD77 pKa = 4.02EE78 pKa = 5.28LYY80 pKa = 10.85LYY82 pKa = 8.18DD83 pKa = 3.94TASGTVLAHH92 pKa = 6.59CPAPAPMRR100 pKa = 11.84DD101 pKa = 3.61FGAAPFKK108 pKa = 10.87DD109 pKa = 4.42GIVLTVMGDD118 pKa = 3.52AGAVAYY124 pKa = 9.7IYY126 pKa = 10.7DD127 pKa = 3.95RR128 pKa = 11.84EE129 pKa = 4.29LKK131 pKa = 10.87LEE133 pKa = 3.99EE134 pKa = 3.98TLALEE139 pKa = 4.6EE140 pKa = 4.83LLSGDD145 pKa = 3.8PVVDD149 pKa = 4.6PGCVAASSDD158 pKa = 3.24GRR160 pKa = 11.84QLAIAGLSALYY171 pKa = 10.34LYY173 pKa = 9.99DD174 pKa = 4.31RR175 pKa = 11.84PAGQLTTLLDD185 pKa = 3.59TAQGVGGASCMATSLNGAAFTPDD208 pKa = 3.61DD209 pKa = 4.1DD210 pKa = 3.91RR211 pKa = 11.84VAFFGDD217 pKa = 3.77GFSAEE222 pKa = 4.46SGNSEE227 pKa = 4.35DD228 pKa = 4.99SAPVWGTIGIDD239 pKa = 3.64GSEE242 pKa = 5.08LEE244 pKa = 4.5LDD246 pKa = 3.36WLEE249 pKa = 4.33SEE251 pKa = 4.95GVEE254 pKa = 4.48EE255 pKa = 4.39MLRR258 pKa = 11.84ADD260 pKa = 3.44GRR262 pKa = 11.84LFFPPAISHH271 pKa = 6.26VDD273 pKa = 3.33GGLPWVDD280 pKa = 3.31AVSGAAHH287 pKa = 7.11RR288 pKa = 11.84LAFSASDD295 pKa = 3.34EE296 pKa = 4.6GGDD299 pKa = 3.65GIYY302 pKa = 10.64VSEE305 pKa = 4.08QGKK308 pKa = 9.27YY309 pKa = 10.02AATAEE314 pKa = 4.38LGDD317 pKa = 3.83VLTVRR322 pKa = 11.84VYY324 pKa = 11.01AVDD327 pKa = 3.42SGEE330 pKa = 4.2LLATEE335 pKa = 4.74TIEE338 pKa = 4.75VDD340 pKa = 2.88DD341 pKa = 3.87SLYY344 pKa = 10.78VGRR347 pKa = 11.84IPQVILLDD355 pKa = 4.09DD356 pKa = 4.11AQAAIVLLGCGIEE369 pKa = 4.51EE370 pKa = 4.62IDD372 pKa = 3.58TAVATFSFGVV382 pKa = 3.31
MM1 pKa = 7.51ALSLLVLGACSAPAPTDD18 pKa = 3.45NSEE21 pKa = 4.0KK22 pKa = 10.58AAPEE26 pKa = 4.08TPAAPLPASDD36 pKa = 4.59VMKK39 pKa = 10.88NDD41 pKa = 5.13DD42 pKa = 3.68PTQTSGFFAFDD53 pKa = 3.05SALFQGKK60 pKa = 7.86LTGCSYY66 pKa = 11.28AQAGTLLVCADD77 pKa = 4.02EE78 pKa = 5.28LYY80 pKa = 10.85LYY82 pKa = 8.18DD83 pKa = 3.94TASGTVLAHH92 pKa = 6.59CPAPAPMRR100 pKa = 11.84DD101 pKa = 3.61FGAAPFKK108 pKa = 10.87DD109 pKa = 4.42GIVLTVMGDD118 pKa = 3.52AGAVAYY124 pKa = 9.7IYY126 pKa = 10.7DD127 pKa = 3.95RR128 pKa = 11.84EE129 pKa = 4.29LKK131 pKa = 10.87LEE133 pKa = 3.99EE134 pKa = 3.98TLALEE139 pKa = 4.6EE140 pKa = 4.83LLSGDD145 pKa = 3.8PVVDD149 pKa = 4.6PGCVAASSDD158 pKa = 3.24GRR160 pKa = 11.84QLAIAGLSALYY171 pKa = 10.34LYY173 pKa = 9.99DD174 pKa = 4.31RR175 pKa = 11.84PAGQLTTLLDD185 pKa = 3.59TAQGVGGASCMATSLNGAAFTPDD208 pKa = 3.61DD209 pKa = 4.1DD210 pKa = 3.91RR211 pKa = 11.84VAFFGDD217 pKa = 3.77GFSAEE222 pKa = 4.46SGNSEE227 pKa = 4.35DD228 pKa = 4.99SAPVWGTIGIDD239 pKa = 3.64GSEE242 pKa = 5.08LEE244 pKa = 4.5LDD246 pKa = 3.36WLEE249 pKa = 4.33SEE251 pKa = 4.95GVEE254 pKa = 4.48EE255 pKa = 4.39MLRR258 pKa = 11.84ADD260 pKa = 3.44GRR262 pKa = 11.84LFFPPAISHH271 pKa = 6.26VDD273 pKa = 3.33GGLPWVDD280 pKa = 3.31AVSGAAHH287 pKa = 7.11RR288 pKa = 11.84LAFSASDD295 pKa = 3.34EE296 pKa = 4.6GGDD299 pKa = 3.65GIYY302 pKa = 10.64VSEE305 pKa = 4.08QGKK308 pKa = 9.27YY309 pKa = 10.02AATAEE314 pKa = 4.38LGDD317 pKa = 3.83VLTVRR322 pKa = 11.84VYY324 pKa = 11.01AVDD327 pKa = 3.42SGEE330 pKa = 4.2LLATEE335 pKa = 4.74TIEE338 pKa = 4.75VDD340 pKa = 2.88DD341 pKa = 3.87SLYY344 pKa = 10.78VGRR347 pKa = 11.84IPQVILLDD355 pKa = 4.09DD356 pKa = 4.11AQAAIVLLGCGIEE369 pKa = 4.51EE370 pKa = 4.62IDD372 pKa = 3.58TAVATFSFGVV382 pKa = 3.31
Molecular weight: 39.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C9NBS4|A0A7C9NBS4_9BACT 4Fe-4S dicluster domain-containing protein OS=Muribaculaceae bacterium Z82 OX=2304548 GN=D1639_09755 PE=4 SV=1
MM1 pKa = 7.75PNGRR5 pKa = 11.84KK6 pKa = 8.9QPASASFRR14 pKa = 11.84RR15 pKa = 11.84TASSPGHH22 pKa = 5.56RR23 pKa = 11.84RR24 pKa = 11.84SRR26 pKa = 11.84SARR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QGRR34 pKa = 11.84SRR36 pKa = 11.84TPAAA40 pKa = 4.06
MM1 pKa = 7.75PNGRR5 pKa = 11.84KK6 pKa = 8.9QPASASFRR14 pKa = 11.84RR15 pKa = 11.84TASSPGHH22 pKa = 5.56RR23 pKa = 11.84RR24 pKa = 11.84SRR26 pKa = 11.84SARR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84QGRR34 pKa = 11.84SRR36 pKa = 11.84TPAAA40 pKa = 4.06
Molecular weight: 4.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
760548 |
27 |
23345 |
343.5 |
37.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.285 ± 0.104 | 1.564 ± 0.048 |
6.14 ± 0.11 | 6.359 ± 0.056 |
3.709 ± 0.043 | 8.657 ± 0.05 |
1.617 ± 0.021 | 4.351 ± 0.055 |
3.483 ± 0.038 | 9.187 ± 0.11 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.603 ± 0.042 | 2.835 ± 0.093 |
4.511 ± 0.033 | 3.167 ± 0.031 |
5.975 ± 0.088 | 6.107 ± 0.048 |
5.181 ± 0.129 | 8.159 ± 0.047 |
1.187 ± 0.029 | 2.924 ± 0.081 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
