
Human gut gokushovirus
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; Gokushovirinae; unclassified Gokushovirinae
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
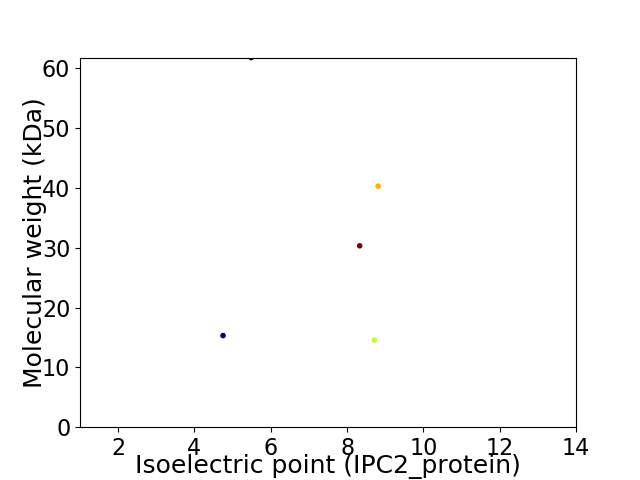
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
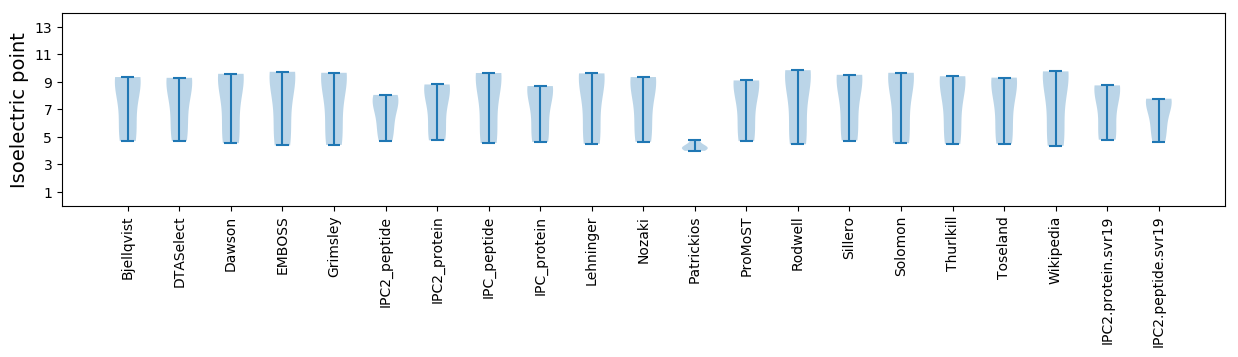
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X9Q0X9|A0A1X9Q0X9_9VIRU Putative VP1 OS=Human gut gokushovirus OX=1986031 PE=3 SV=1
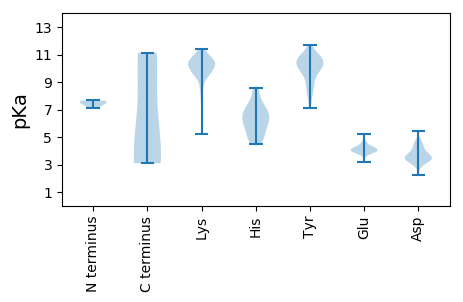
MM1 pKa = 7.37NKK3 pKa = 9.69RR4 pKa = 11.84YY5 pKa = 10.12EE6 pKa = 4.15EE7 pKa = 4.1GRR9 pKa = 11.84EE10 pKa = 3.91PFFSEE15 pKa = 3.91SGEE18 pKa = 4.2KK19 pKa = 9.89FRR21 pKa = 11.84KK22 pKa = 8.98QYY24 pKa = 10.61VWTKK28 pKa = 10.67DD29 pKa = 3.35EE30 pKa = 4.07QGQEE34 pKa = 4.25VLQEE38 pKa = 3.91TAPIDD43 pKa = 3.37IQQEE47 pKa = 3.96IEE49 pKa = 4.6SYY51 pKa = 11.36AEE53 pKa = 3.78EE54 pKa = 4.7CDD56 pKa = 2.99IKK58 pKa = 11.3SIVRR62 pKa = 11.84KK63 pKa = 10.09ASFDD67 pKa = 3.67PQFLKK72 pKa = 10.93SLSEE76 pKa = 3.91GAMTGEE82 pKa = 4.18EE83 pKa = 3.83VDD85 pKa = 3.21ITEE88 pKa = 4.85FPQNIHH94 pKa = 6.34EE95 pKa = 4.31YY96 pKa = 10.49HH97 pKa = 6.87RR98 pKa = 11.84MIATAQANAMKK109 pKa = 10.67LEE111 pKa = 4.05EE112 pKa = 4.23LQKK115 pKa = 10.92KK116 pKa = 9.07AAEE119 pKa = 4.03EE120 pKa = 3.95AKK122 pKa = 9.99TEE124 pKa = 4.19SEE126 pKa = 4.07TKK128 pKa = 9.92EE129 pKa = 3.92AEE131 pKa = 4.18KK132 pKa = 10.99
MM1 pKa = 7.37NKK3 pKa = 9.69RR4 pKa = 11.84YY5 pKa = 10.12EE6 pKa = 4.15EE7 pKa = 4.1GRR9 pKa = 11.84EE10 pKa = 3.91PFFSEE15 pKa = 3.91SGEE18 pKa = 4.2KK19 pKa = 9.89FRR21 pKa = 11.84KK22 pKa = 8.98QYY24 pKa = 10.61VWTKK28 pKa = 10.67DD29 pKa = 3.35EE30 pKa = 4.07QGQEE34 pKa = 4.25VLQEE38 pKa = 3.91TAPIDD43 pKa = 3.37IQQEE47 pKa = 3.96IEE49 pKa = 4.6SYY51 pKa = 11.36AEE53 pKa = 3.78EE54 pKa = 4.7CDD56 pKa = 2.99IKK58 pKa = 11.3SIVRR62 pKa = 11.84KK63 pKa = 10.09ASFDD67 pKa = 3.67PQFLKK72 pKa = 10.93SLSEE76 pKa = 3.91GAMTGEE82 pKa = 4.18EE83 pKa = 3.83VDD85 pKa = 3.21ITEE88 pKa = 4.85FPQNIHH94 pKa = 6.34EE95 pKa = 4.31YY96 pKa = 10.49HH97 pKa = 6.87RR98 pKa = 11.84MIATAQANAMKK109 pKa = 10.67LEE111 pKa = 4.05EE112 pKa = 4.23LQKK115 pKa = 10.92KK116 pKa = 9.07AAEE119 pKa = 4.03EE120 pKa = 3.95AKK122 pKa = 9.99TEE124 pKa = 4.19SEE126 pKa = 4.07TKK128 pKa = 9.92EE129 pKa = 3.92AEE131 pKa = 4.18KK132 pKa = 10.99
Molecular weight: 15.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X9Q0X5|A0A1X9Q0X5_9VIRU Peptidase M15 OS=Human gut gokushovirus OX=1986031 PE=4 SV=1
MM1 pKa = 7.48SCYY4 pKa = 10.22KK5 pKa = 10.36PLIRR9 pKa = 11.84LYY11 pKa = 10.75NPNDD15 pKa = 3.53KK16 pKa = 10.36EE17 pKa = 3.92ISGRR21 pKa = 11.84VYY23 pKa = 10.31SLSRR27 pKa = 11.84FSQLAGKK34 pKa = 7.37QLKK37 pKa = 10.34YY38 pKa = 10.6EE39 pKa = 4.03DD40 pKa = 4.56LMYY43 pKa = 10.69KK44 pKa = 9.86KK45 pKa = 10.34NVMLIPCGQCIGCRR59 pKa = 11.84IRR61 pKa = 11.84QRR63 pKa = 11.84EE64 pKa = 4.0DD65 pKa = 2.26WTTRR69 pKa = 11.84IEE71 pKa = 4.51LEE73 pKa = 3.75ARR75 pKa = 11.84DD76 pKa = 4.0YY77 pKa = 11.23PKK79 pKa = 10.6EE80 pKa = 3.92QVWFITLTYY89 pKa = 10.91DD90 pKa = 3.88DD91 pKa = 4.65DD92 pKa = 4.54HH93 pKa = 8.55VPGMIINTGEE103 pKa = 3.91IMRR106 pKa = 11.84KK107 pKa = 7.5VQYY110 pKa = 9.75VWRR113 pKa = 11.84PGEE116 pKa = 3.89KK117 pKa = 9.53RR118 pKa = 11.84PEE120 pKa = 4.07SVQTLLYY127 pKa = 10.51TDD129 pKa = 3.04IQKK132 pKa = 10.17FLKK135 pKa = 10.14RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 9.69AYY141 pKa = 9.94GGKK144 pKa = 9.63LRR146 pKa = 11.84YY147 pKa = 8.78FVAGEE152 pKa = 4.04YY153 pKa = 10.92GEE155 pKa = 4.36QTARR159 pKa = 11.84PHH161 pKa = 4.93YY162 pKa = 10.8HH163 pKa = 6.27MILYY167 pKa = 8.99GWQPTDD173 pKa = 4.23LEE175 pKa = 4.28HH176 pKa = 7.5LYY178 pKa = 10.5KK179 pKa = 10.14IQHH182 pKa = 6.03NGYY185 pKa = 7.86FTSKK189 pKa = 9.62WLEE192 pKa = 4.19DD193 pKa = 3.66LWGMGQIQIAQAVPEE208 pKa = 4.37TYY210 pKa = 9.7RR211 pKa = 11.84YY212 pKa = 8.61VAGYY216 pKa = 7.14VTKK219 pKa = 10.66KK220 pKa = 9.59MYY222 pKa = 10.72EE223 pKa = 3.63IDD225 pKa = 3.75GQKK228 pKa = 10.9ANAYY232 pKa = 9.87YY233 pKa = 10.54EE234 pKa = 4.35LGQTKK239 pKa = 9.87PFACMSLKK247 pKa = 10.53PGLGDD252 pKa = 4.31HH253 pKa = 7.19YY254 pKa = 10.28YY255 pKa = 10.16QEE257 pKa = 4.59HH258 pKa = 6.52KK259 pKa = 11.28AEE261 pKa = 4.14IWRR264 pKa = 11.84QGYY267 pKa = 7.72IQCTNGKK274 pKa = 8.15HH275 pKa = 5.09AQIPRR280 pKa = 11.84YY281 pKa = 7.47YY282 pKa = 9.34EE283 pKa = 3.81KK284 pKa = 10.57MMEE287 pKa = 4.21AEE289 pKa = 4.03NPQRR293 pKa = 11.84LWKK296 pKa = 10.3IKK298 pKa = 8.01QNRR301 pKa = 11.84QAAAIAEE308 pKa = 4.05NRR310 pKa = 11.84LKK312 pKa = 11.21YY313 pKa = 10.68EE314 pKa = 3.84NTDD317 pKa = 3.45FAEE320 pKa = 4.0QCKK323 pKa = 9.23TKK325 pKa = 10.54EE326 pKa = 3.92RR327 pKa = 11.84VIKK330 pKa = 10.55KK331 pKa = 8.58QMKK334 pKa = 10.18KK335 pKa = 10.39KK336 pKa = 9.42GTLL339 pKa = 3.25
MM1 pKa = 7.48SCYY4 pKa = 10.22KK5 pKa = 10.36PLIRR9 pKa = 11.84LYY11 pKa = 10.75NPNDD15 pKa = 3.53KK16 pKa = 10.36EE17 pKa = 3.92ISGRR21 pKa = 11.84VYY23 pKa = 10.31SLSRR27 pKa = 11.84FSQLAGKK34 pKa = 7.37QLKK37 pKa = 10.34YY38 pKa = 10.6EE39 pKa = 4.03DD40 pKa = 4.56LMYY43 pKa = 10.69KK44 pKa = 9.86KK45 pKa = 10.34NVMLIPCGQCIGCRR59 pKa = 11.84IRR61 pKa = 11.84QRR63 pKa = 11.84EE64 pKa = 4.0DD65 pKa = 2.26WTTRR69 pKa = 11.84IEE71 pKa = 4.51LEE73 pKa = 3.75ARR75 pKa = 11.84DD76 pKa = 4.0YY77 pKa = 11.23PKK79 pKa = 10.6EE80 pKa = 3.92QVWFITLTYY89 pKa = 10.91DD90 pKa = 3.88DD91 pKa = 4.65DD92 pKa = 4.54HH93 pKa = 8.55VPGMIINTGEE103 pKa = 3.91IMRR106 pKa = 11.84KK107 pKa = 7.5VQYY110 pKa = 9.75VWRR113 pKa = 11.84PGEE116 pKa = 3.89KK117 pKa = 9.53RR118 pKa = 11.84PEE120 pKa = 4.07SVQTLLYY127 pKa = 10.51TDD129 pKa = 3.04IQKK132 pKa = 10.17FLKK135 pKa = 10.14RR136 pKa = 11.84LRR138 pKa = 11.84KK139 pKa = 9.69AYY141 pKa = 9.94GGKK144 pKa = 9.63LRR146 pKa = 11.84YY147 pKa = 8.78FVAGEE152 pKa = 4.04YY153 pKa = 10.92GEE155 pKa = 4.36QTARR159 pKa = 11.84PHH161 pKa = 4.93YY162 pKa = 10.8HH163 pKa = 6.27MILYY167 pKa = 8.99GWQPTDD173 pKa = 4.23LEE175 pKa = 4.28HH176 pKa = 7.5LYY178 pKa = 10.5KK179 pKa = 10.14IQHH182 pKa = 6.03NGYY185 pKa = 7.86FTSKK189 pKa = 9.62WLEE192 pKa = 4.19DD193 pKa = 3.66LWGMGQIQIAQAVPEE208 pKa = 4.37TYY210 pKa = 9.7RR211 pKa = 11.84YY212 pKa = 8.61VAGYY216 pKa = 7.14VTKK219 pKa = 10.66KK220 pKa = 9.59MYY222 pKa = 10.72EE223 pKa = 3.63IDD225 pKa = 3.75GQKK228 pKa = 10.9ANAYY232 pKa = 9.87YY233 pKa = 10.54EE234 pKa = 4.35LGQTKK239 pKa = 9.87PFACMSLKK247 pKa = 10.53PGLGDD252 pKa = 4.31HH253 pKa = 7.19YY254 pKa = 10.28YY255 pKa = 10.16QEE257 pKa = 4.59HH258 pKa = 6.52KK259 pKa = 11.28AEE261 pKa = 4.14IWRR264 pKa = 11.84QGYY267 pKa = 7.72IQCTNGKK274 pKa = 8.15HH275 pKa = 5.09AQIPRR280 pKa = 11.84YY281 pKa = 7.47YY282 pKa = 9.34EE283 pKa = 3.81KK284 pKa = 10.57MMEE287 pKa = 4.21AEE289 pKa = 4.03NPQRR293 pKa = 11.84LWKK296 pKa = 10.3IKK298 pKa = 8.01QNRR301 pKa = 11.84QAAAIAEE308 pKa = 4.05NRR310 pKa = 11.84LKK312 pKa = 11.21YY313 pKa = 10.68EE314 pKa = 3.84NTDD317 pKa = 3.45FAEE320 pKa = 4.0QCKK323 pKa = 9.23TKK325 pKa = 10.54EE326 pKa = 3.92RR327 pKa = 11.84VIKK330 pKa = 10.55KK331 pKa = 8.58QMKK334 pKa = 10.18KK335 pKa = 10.39KK336 pKa = 9.42GTLL339 pKa = 3.25
Molecular weight: 40.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1434 |
130 |
544 |
286.8 |
32.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.741 ± 0.974 | 0.976 ± 0.363 |
4.533 ± 0.509 | 7.322 ± 1.556 |
3.347 ± 0.629 | 8.159 ± 1.384 |
1.743 ± 0.29 | 5.649 ± 0.846 |
7.462 ± 1.096 | 5.509 ± 0.602 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.208 ± 0.375 | 5.021 ± 0.774 |
3.975 ± 0.678 | 5.858 ± 1.176 |
4.742 ± 0.854 | 5.788 ± 1.711 |
6.206 ± 0.634 | 5.439 ± 1.17 |
1.674 ± 0.254 | 5.649 ± 0.849 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
